8HUJ
 
 | | Cryo-EM structure of the J-K-St region of EMCV IRES in complex with eIF4G-HEAT1 and eIF4A | | 分子名称: | Eukaryotic initiation factor 4A-I, Eukaryotic translation initiation factor 4 gamma 1, IRES RNA (J-K-St), ... | | 著者 | Suzuki, H, Fujiyoshi, Y, Imai, S, Shimada, I. | | 登録日 | 2022-12-24 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | ELECTRON MICROSCOPY (3.76 Å) | | 主引用文献 | Dynamically regulated two-site interaction of viral RNA to capture host translation initiation factor.
Nat Commun, 14, 2023
|
|
7F8J
 
 | | Cryo-EM structure of human pannexin-1 in a nanodisc | | 分子名称: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | 著者 | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | 登録日 | 2021-07-02 | | 公開日 | 2022-01-26 | | 最終更新日 | 2022-02-23 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7F8O
 
 | | Cryo-EM structure of the C-terminal deletion mutant of human PANX1 in a nanodisc | | 分子名称: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | 著者 | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | 登録日 | 2021-07-02 | | 公開日 | 2022-01-26 | | 最終更新日 | 2022-02-23 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7F8N
 
 | | Human pannexin-1 showing a conformational change in the N-terminal domain and blocked pore | | 分子名称: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | 著者 | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | 登録日 | 2021-07-02 | | 公開日 | 2022-01-26 | | 最終更新日 | 2022-02-23 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
1K5O
 
 | | CPI-17(35-120) deletion mutant | | 分子名称: | CPI-17 | | 著者 | Ohki, S, Eto, M, Kariya, E, Hayano, T, Hayashi, Y, Yazawa, M, Brautigan, D, Kainosho, M. | | 登録日 | 2001-10-11 | | 公開日 | 2002-10-11 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of the Myosin Phosphatase Inhibitor Protein CPI-17 Shows Phosphorylation-induced Conformational Changes Responsible for Activation
J.Mol.Biol., 314, 2001
|
|
1KRL
 
 | | Crystal Structure of Racemic DL-monellin in P-1 | | 分子名称: | MONELLIN, CHAIN A, CHAIN B | | 著者 | Hung, L.W, Kohmura, M, Ariyoshi, Y, Kim, S.H. | | 登録日 | 2002-01-10 | | 公開日 | 2002-02-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural differences in D and L-monellin in the crystals of racemic mixture.
J.Mol.Biol., 285, 1999
|
|
4MSP
 
 | | Crystal structure of human peptidyl-prolyl cis-trans isomerase FKBP22 (aka FKBP14) containing two EF-hand motifs | | 分子名称: | CALCIUM ION, PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase FKBP14, ... | | 著者 | Boudko, S.P, Ishikawa, Y, Bachinger, H.P. | | 登録日 | 2013-09-18 | | 公開日 | 2013-12-25 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of human peptidyl-prolyl cis-trans isomerase FKBP22 containing two EF-hand motifs.
Protein Sci., 23, 2014
|
|
1QQI
 
 | | SOLUTION STRUCTURE OF THE DNA-BINDING AND TRANSACTIVATION DOMAIN OF PHOB FROM ESCHERICHIA COLI | | 分子名称: | PHOSPHATE REGULON TRANSCRIPTIONAL REGULATORY PROTEIN PHOB | | 著者 | Okamura, H, Hanaoka, S, Nagadoi, A, Makino, K, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 1999-06-07 | | 公開日 | 2000-06-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural comparison of the PhoB and OmpR DNA-binding/transactivation domains and the arrangement of PhoB molecules on the phosphate box.
J.Mol.Biol., 295, 2000
|
|
1MHW
 
 | | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides | | 分子名称: | 4-biphenylacetyl-Cys-(D)Arg-Tyr-N-(2-phenylethyl) amide, Cathepsin L | | 著者 | Chowdhury, S, Sivaraman, J, Wang, J, Devanathan, G, Lachance, P, Qi, H, Menard, R, Lefebvre, J, Konishi, Y, Cygler, M, Sulea, T, Purisima, E.O. | | 登録日 | 2002-08-21 | | 公開日 | 2002-12-11 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides
J.Med.Chem., 45, 2002
|
|
2ZCZ
 
 | | Crystal structures and thermostability of mutant TRAP3 A7 (ENGINEERED TRAP) | | 分子名称: | TRYPTOPHAN, Transcription attenuation protein mtrB | | 著者 | Watanabe, M, Mishima, Y, Yamashita, I, Park, S.Y, Tame, J.R.H, Heddle, J.G. | | 登録日 | 2007-11-15 | | 公開日 | 2008-04-29 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Intersubunit linker length as a modifier of protein stability: crystal structures and thermostability of mutant TRAP.
Protein Sci., 17, 2008
|
|
2PA2
 
 | | Crystal structure of human Ribosomal protein L10 core domain | | 分子名称: | 60S ribosomal protein L10, POTASSIUM ION | | 著者 | Nishimura, M, Kaminishi, T, Takemoto, C, Kawazoe, M, Yoshida, T, Tanaka, A, Sugano, S, Shirouzu, M, Ohkubo, T, Yokoyama, S, Kobayashi, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-03-27 | | 公開日 | 2008-03-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of Human Ribosomal Protein L10 Core Domain Reveals Eukaryote-Specific Motifs in Addition to the Conserved Fold
J.Mol.Biol., 377, 2008
|
|
8J7R
 
 | | Cryo-EM structure of the J-K-St region of EMCV IRES in complex with eIF4G-HEAT1 and eIF4A (J-K-St/eIF4G focused) | | 分子名称: | Eukaryotic translation initiation factor 4 gamma 1, IRES RNA (J-K-St), MAGNESIUM ION | | 著者 | Suzuki, H, Fujiyoshi, Y, Imai, S, Shimada, I. | | 登録日 | 2023-04-28 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Dynamically regulated two-site interaction of viral RNA to capture host translation initiation factor.
Nat Commun, 14, 2023
|
|
2ZD0
 
 | | Crystal structures and thermostability of mutant TRAP3 A5 (ENGINEERED TRAP) | | 分子名称: | TRYPTOPHAN, Transcription attenuation protein mtrB | | 著者 | Watanabe, M, Mishima, Y, Yamashita, I, Park, S.Y, Tame, J.R.H, Heddle, J.G. | | 登録日 | 2007-11-15 | | 公開日 | 2008-04-29 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Intersubunit linker length as a modifier of protein stability: crystal structures and thermostability of mutant TRAP.
Protein Sci., 17, 2008
|
|
2PCR
 
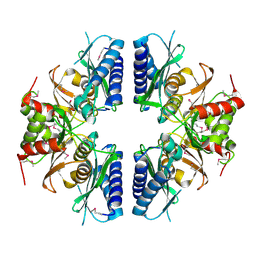 | | Crystal structure of Myo-inositol-1(or 4)-monophosphatase (aq_1983) from Aquifex Aeolicus VF5 | | 分子名称: | Inositol-1-monophosphatase | | 著者 | Jeyakanthan, J, Gayathri, D, Velmurugan, D, Agari, Y, Bessho, Y, Ellis, M.J, Antonyuk, S.V, Strange, R.W, Hasnain, S.S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-03-30 | | 公開日 | 2007-10-02 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of Myo-inositol-1(or 4)-monophosphatase (aq_1983) from Aquifex Aeolicus VF5
To be Published
|
|
1RNF
 
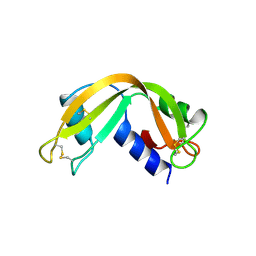 | | X-RAY CRYSTAL STRUCTURE OF UNLIGANDED HUMAN RIBONUCLEASE 4 | | 分子名称: | PROTEIN (RIBONUCLEASE 4) | | 著者 | Terzyan, S.S, Peracaula, R, De Llorens, R, Tsushima, Y, Yamada, H, Seno, M, Gomis-Rueth, F.X, Coll, M. | | 登録日 | 1998-10-29 | | 公開日 | 1999-10-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The three-dimensional structure of human RNase 4, unliganded and complexed with d(Up), reveals the basis for its uridine selectivity.
J.Mol.Biol., 285, 1999
|
|
2MXU
 
 | | 42-Residue Beta Amyloid Fibril | | 分子名称: | Amyloid beta A4 protein | | 著者 | Xiao, Y, Ma, B, McElheny, D, Parthasarathy, S, Long, F, Hoshi, M, Nussinov, R, Ishii, Y. | | 登録日 | 2015-01-14 | | 公開日 | 2015-05-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | A beta (1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer's disease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
2OMD
 
 | | Crystal structure of molybdopterin converting factor subunit 2 (aq_2181) from aquifex aeolicus VF5 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-01-22 | | 公開日 | 2008-01-29 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of molybdopterin converting factor subunit 2 (aq_2181) from aquifex aeolicus VF5
To be Published
|
|
2PBP
 
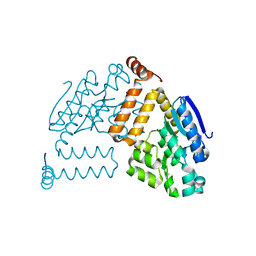 | | Crystal structure of ENOYL-CoA hydrates subunit I (gk_2039) from geobacillus kaustophilus HTA426 | | 分子名称: | Enoyl-CoA hydratase subunit I | | 著者 | Jeyakanthan, J, Kanaujia, S.P, Vasuki, R.C, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-03-29 | | 公開日 | 2007-10-02 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of ENOYL-CoA hydrates subunit I (gk_2039) from geobacillus kaustophilus HTA426
To be Published
|
|
2PBQ
 
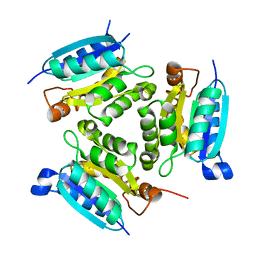 | | Crystal structure of molybdenum cofactor biosynthesis (aq_061) From aquifex aeolicus VF5 | | 分子名称: | Molybdenum cofactor biosynthesis MOG | | 著者 | Jeyakanthan, J, Mahesh, S, Kanaujia, S.P, Ramakumar, S, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-03-29 | | 公開日 | 2007-10-02 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of molybdenum cofactor biosynthesis (aq_061) from aquifex aeolicus VF5
to be published
|
|
7YH6
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-8 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, NIV-8 Fab light chain, ... | | 著者 | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | 登録日 | 2022-07-12 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
3U2J
 
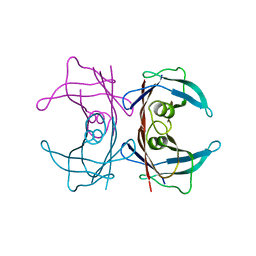 | | Neutron crystal structure of human Transthyretin | | 分子名称: | Transthyretin | | 著者 | Yokoyama, T, Mizuguchi, M, Nabeshima, Y, Kusaka, K, Yamada, T, Hosoya, T, Ohhara, T, Kurihara, K, Tomoyori, K, Tanaka, I, Niimura, N. | | 登録日 | 2011-10-03 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-11-01 | | 実験手法 | NEUTRON DIFFRACTION (2 Å) | | 主引用文献 | Hydrogen-bond network and pH sensitivity in transthyretin: Neutron crystal structure of human transthyretin
J.Struct.Biol., 177, 2012
|
|
7YH7
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-8 (state 2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, ... | | 著者 | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | 登録日 | 2022-07-13 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
3U2I
 
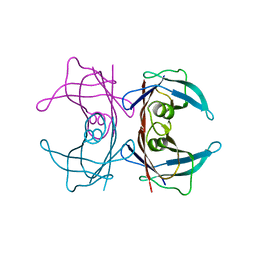 | | X-ray crystal structure of human Transthyretin at room temperature | | 分子名称: | Transthyretin | | 著者 | Yokoyama, T, Mizuguchi, M, Nabeshima, Y, Kusaka, K, Yamada, T, Hosoya, T, Ohhara, T, Kurihara, K, Tomoyori, K, Tanaka, I, Niimura, N. | | 登録日 | 2011-10-03 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Hydrogen-bond network and pH sensitivity in transthyretin: Neutron crystal structure of human transthyretin
J.Struct.Biol., 177, 2012
|
|
2PQ0
 
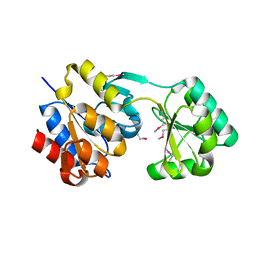 | | Crystal structure of Hyopthetical protein (gk_1056) from geobacillus Kaustophilus HTA426 | | 分子名称: | Hypothetical conserved protein GK1056 | | 著者 | Kanaujia, S.P, Jeyakanthan, J, Kavyashree, M, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-05-01 | | 公開日 | 2008-05-06 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of Hyopthetical protein (gk_1056) from geobacillus Kaustophilus HTA426
To be Published
|
|
2PCQ
 
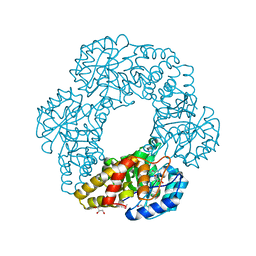 | | Crystal structure of putative dihydrodipicolinate synthase (TTHA0737) from Thermus Thermophilus HB8 | | 分子名称: | GLYCEROL, POTASSIUM ION, Putative dihydrodipicolinate synthase | | 著者 | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Kitamura, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-03-30 | | 公開日 | 2007-10-02 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of putative dihydrodipicolinate synthase (TTHA0737) from Thermus Thermophilus HB8
To be Published
|
|
