4FCE
 
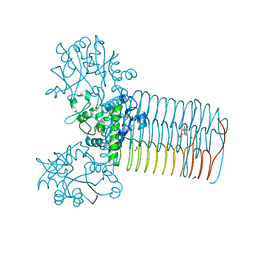 | | Crystal structure of Yersinia pestis GlmU in complex with alpha-D-glucosamine 1-phosphate (GP1) | | 分子名称: | 1,2-ETHANEDIOL, 2-amino-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, Bifunctional protein GlmU, ... | | 著者 | Nocek, B, Kuhn, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-05-24 | | 公開日 | 2012-07-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.955 Å) | | 主引用文献 | Crystal structure of Yersinia pestis GlmU in complex with alpha-D-glucosamine 1-phosphate (GP1)
To be Published
|
|
4DYU
 
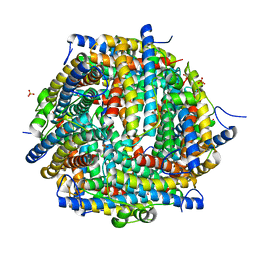 | | The crystal structure of DNA starvation/stationary phase protection protein Dps from Yersinia pestis KIM 10 | | 分子名称: | DNA protection during starvation protein, SULFATE ION, ZINC ION | | 著者 | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-02-29 | | 公開日 | 2012-03-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | The crystal structure of DNA starvation/stationary phase protection protein Dps from Yersinia pestis KIM 10
To be Published
|
|
4E16
 
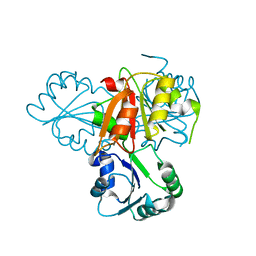 | | Precorrin-4 C(11)-methyltransferase from Clostridium difficile | | 分子名称: | precorrin-4 C(11)-methyltransferase | | 著者 | Osipiuk, J, Nocek, B, Makowska-Grzyska, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-03-05 | | 公開日 | 2012-03-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Precorrin-4 C(11)-methyltransferase from Clostridium difficile
To be Published
|
|
4QMK
 
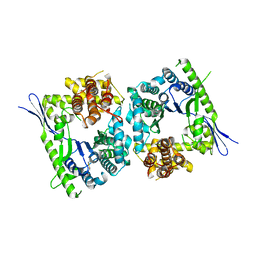 | | Crystal structure of type III effector protein ExoU (exoU) | | 分子名称: | BETA-MERCAPTOETHANOL, Type III secretion system effector protein ExoU | | 著者 | Halavaty, A.S, Tyson, G.H, Zhang, A, Hauser, A.R, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-06-16 | | 公開日 | 2014-12-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A Novel Phosphatidylinositol 4,5-Bisphosphate Binding Domain Mediates Plasma Membrane Localization of ExoU and Other Patatin-like Phospholipases.
J.Biol.Chem., 290, 2015
|
|
4QJ1
 
 | | Co-crystal structure of the catalytic domain of the inosine monophosphate dehydrogenase from Cryptosporidium parvum with inhibitor N109 | | 分子名称: | 1,2-ETHANEDIOL, ACETIC ACID, FORMIC ACID, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-06-03 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.415 Å) | | 主引用文献 | Co-crystal structure of the catalytic domain of the inosine monophosphate dehydrogenase from Cryptosporidium parvum with inhibitor N109
To be Published
|
|
4RV8
 
 | | Co-Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Cryptosporidium parvum and the inhibitor p131 | | 分子名称: | 1-(2-{3-[(1E)-N-(2-aminoethoxy)ethanimidoyl]phenyl}propan-2-yl)-3-(4-chloro-3-nitrophenyl)urea, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-11-25 | | 公開日 | 2014-12-31 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.053 Å) | | 主引用文献 | Structure of Cryptosporidium IMP dehydrogenase bound to an inhibitor with in vivo antiparasitic activity.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
4RWR
 
 | | 2.1 Angstrom Crystal Structure of Stage II Sporulation Protein D from Bacillus anthracis | | 分子名称: | Stage II sporulation protein D | | 著者 | Minasov, G, Wawrzak, Z, Nocadello, S, Shuvalova, L, Dubrovska, I, Flores, K, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-12-05 | | 公開日 | 2014-12-17 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structures of the SpoIID Lytic Transglycosylases Essential for Bacterial Sporulation.
J.Biol.Chem., 291, 2016
|
|
4S12
 
 | | 1.55 Angstrom Crystal Structure of N-acetylmuramic acid 6-phosphate Etherase from Yersinia enterocolitica. | | 分子名称: | DI(HYDROXYETHYL)ETHER, N-acetylmuramic acid 6-phosphate etherase, SULFATE ION | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Flores, K, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-01-07 | | 公開日 | 2015-01-21 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | 1.55 Angstrom Crystal Structure of N-acetylmuramic acid 6-phosphate Etherase from Yersinia enterocolitica.
TO BE PUBLISHED
|
|
4R7Q
 
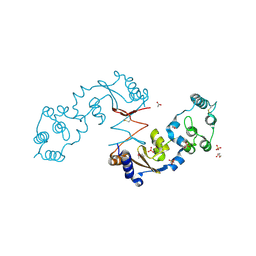 | | The structure of a sensor domain of a histidine kinase from Vibrio cholerae O1 biovar eltor str. N16961 | | 分子名称: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | 著者 | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-08-28 | | 公開日 | 2014-09-10 | | 最終更新日 | 2022-11-16 | | 実験手法 | X-RAY DIFFRACTION (1.981 Å) | | 主引用文献 | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 203, 2021
|
|
4R9M
 
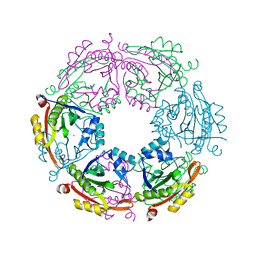 | | Crystal structure of spermidine N-acetyltransferase from Escherichia coli | | 分子名称: | MAGNESIUM ION, Spermidine N(1)-acetyltransferase | | 著者 | Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Grimshaw, S, Wolfe, A.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-09-05 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Analysis of crystalline and solution states of ligand-free spermidine N-acetyltransferase (SpeG) from Escherichia coli.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4FO4
 
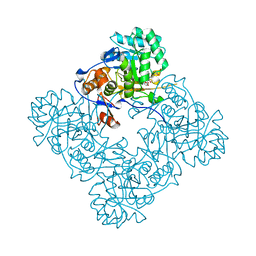 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, complexed with IMP and mycophenolic acid | | 分子名称: | INOSINIC ACID, Inosine 5'-monophosphate dehydrogenase, MYCOPHENOLIC ACID, ... | | 著者 | Osipiuk, J, Maltseva, N, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-06-20 | | 公開日 | 2012-07-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, complexed with IMP and mycophenolic acid.
To be Published
|
|
4OAK
 
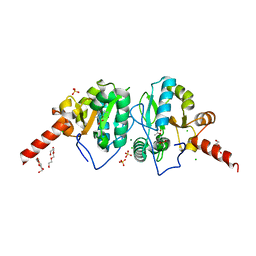 | | Crystal structure of vancomycin resistance D,D-dipeptidase/D,D-pentapeptidase VanXYc D59S mutant in complex with D-Alanine-D-Alanine and copper (II) | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, COPPER (II) ION, ... | | 著者 | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-01-04 | | 公開日 | 2014-01-15 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4FCA
 
 | | The crystal structure of a functionally unknown conserved protein from Bacillus anthracis str. Ames. | | 分子名称: | Conserved domain protein, IMIDAZOLE, NICKEL (II) ION | | 著者 | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-05-24 | | 公開日 | 2012-06-06 | | 実験手法 | X-RAY DIFFRACTION (2.055 Å) | | 主引用文献 | The crystal structure of a functionally unknown conserved protein from Bacillus anthracis str. Ames.
To be Published
|
|
4OFX
 
 | | Crystal Structure of a Putative Cystathionine beta-Synthase from Coxiella burnetii | | 分子名称: | Cystathionine beta-synthase, SODIUM ION | | 著者 | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-01-15 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Crystal Structure of a Putative Cystathionine beta-Synthase from Coxiella burnetii
To be Published
|
|
4OIG
 
 | |
4RN7
 
 | | The crystal structure of N-acetylmuramoyl-L-alanine amidase from Clostridium difficile 630 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMIC ACID, GLYCEROL, ... | | 著者 | Tan, K, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-10-23 | | 公開日 | 2014-11-05 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.717 Å) | | 主引用文献 | The crystal structure of N-acetylmuramoyl-L-alanine amidase from Clostridium difficile 630
To be Published
|
|
4FEZ
 
 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | 著者 | Osipiuk, J, Maltseva, N, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-05-30 | | 公開日 | 2012-06-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant.
To be Published
|
|
4FG0
 
 | |
4FFE
 
 | | The structure of cowpox virus CPXV018 (OMCP) | | 分子名称: | CPXV018 protein | | 著者 | Lazear, E, Peterson, L.W, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-06-01 | | 公開日 | 2012-06-20 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal Structure of the Cowpox Virus-Encoded NKG2D Ligand OMCP.
J.Virol., 87, 2013
|
|
7U5T
 
 | |
7U5S
 
 | |
7U5U
 
 | |
4HKJ
 
 | |
4GUI
 
 | | 1.78 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) in Complex with Quinate | | 分子名称: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, 3-dehydroquinate dehydratase, NICKEL (II) ION | | 著者 | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-08-29 | | 公開日 | 2012-09-12 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Crystal structures of type I dehydroquinate dehydratase in complex with quinate and shikimate suggest a novel mechanism of schiff base formation.
Biochemistry, 53, 2014
|
|
4GUF
 
 | | 1.5 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) E86A Mutant | | 分子名称: | 3-dehydroquinate dehydratase, CHLORIDE ION | | 著者 | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-08-29 | | 公開日 | 2012-09-12 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Reassessing the type I dehydroquinate dehydratase catalytic triad: Kinetic and structural studies of Glu86 mutants.
Protein Sci., 22, 2013
|
|
