8FI2
 
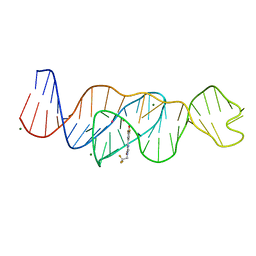 | | Structure of Lettuce C20T bound to DFHBI-1T | | 分子名称: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | 著者 | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | 登録日 | 2022-12-15 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
8FI1
 
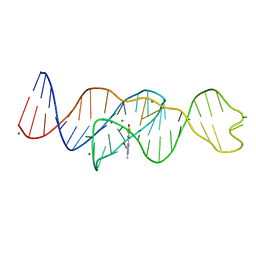 | | Structure of Lettuce C20G bound to DFHO | | 分子名称: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | 著者 | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | 登録日 | 2022-12-15 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
8FI0
 
 | |
8FI7
 
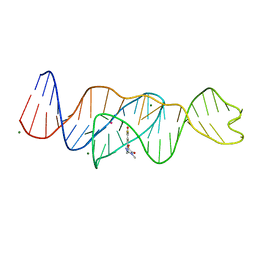 | | Structure of Lettuce C20T bound to DFHO | | 分子名称: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | 著者 | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | 登録日 | 2022-12-15 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
8FHX
 
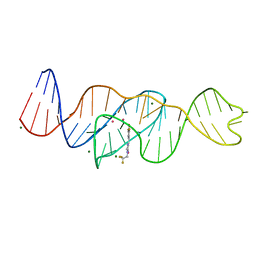 | | Structure of Lettuce aptamer bound to DFHBI-1T | | 分子名称: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | 著者 | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | 登録日 | 2022-12-15 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
8FIQ
 
 | |
8FVT
 
 | |
8FIH
 
 | |
8FIT
 
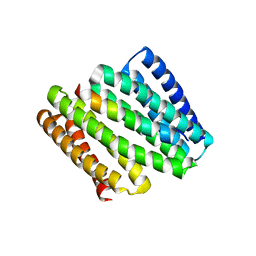 | |
8FIN
 
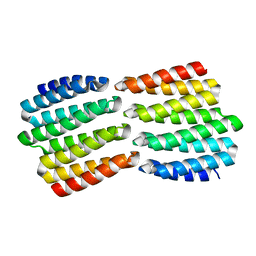 | |
8FWF
 
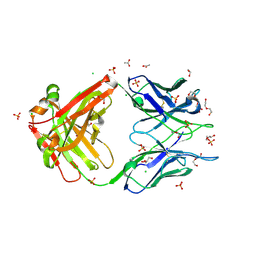 | | Crystal structure of Apo form Fab235 | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Tan, K, Kim, M, Reinherz, E.L. | | 登録日 | 2023-01-21 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
8FXJ
 
 | | Crystal structure of Fab460 | | 分子名称: | ACETATE ION, CHLORIDE ION, Fab460, ... | | 著者 | Tan, K, Kim, M, Reinherz, E.L. | | 登録日 | 2023-01-24 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
8FYM
 
 | | Crystal structure of Fab235 in complex with MPER peptide | | 分子名称: | ALA-SER-LEU-TRP-ASN-TRP-PHE-ASN-ILE-THR-ASN-TRP-LEU-TRP-TYR-ILE-LYS-LYS-LYS, CHLORIDE ION, Fab235, ... | | 著者 | Tan, K, Kim, M, Reinherz, E.L. | | 登録日 | 2023-01-26 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
8FZ2
 
 | | Crystal structure of Fab460 in complex with MPER peptide | | 分子名称: | Fab460, H chain, L chain, ... | | 著者 | Tan, K, Kim, M, Reinherz, E.L. | | 登録日 | 2023-01-27 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
5FGD
 
 | |
5FOL
 
 | | Crystal structure of the Cryptosporidium muris cytosolic leucyl-tRNA synthetase editing domain complex with a post-transfer editing analogue of isoeucine (Ile2AA) | | 分子名称: | 2'-(L-ISOLEUCYL)AMINO-2'-DEOXYADENOSINE, LEUCYL-TRNA SYNTHETASE, PHOSPHATE ION | | 著者 | Palencia, A, Liu, R.J, Lukarska, M, Gut, J, Bougdour, A, Touquet, B, Wang, E.D, Alley, M.R.K, Rosenthal, P.J, Hakimi, M.A, Cusack, S. | | 登録日 | 2015-11-24 | | 公開日 | 2016-08-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Cryptosporidium and Toxoplasma Parasites are Inhibited by a Benzoxaborole Targeting Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5FOM
 
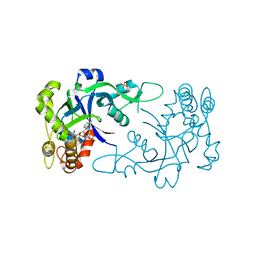 | | Crystal structure of the Cryptosporidium muris cytosolic leucyl-tRNA synthetase editing domain complex with the adduct AMP-AN6426 | | 分子名称: | 4-Chloro-3-aminomethyl-7-[ethoxy]-3H-benzo[C][1,2]oxaborol-1-ol modified adenosine, LEUCYL-TRNA SYNTHETASE, PHOSPHATE ION | | 著者 | Palencia, A, Liu, R.J, Lukarska, M, Gut, J, Bougdour, A, Touquet, B, Wang, E.D, Alley, M.R.K, Rosenthal, P.J, Hakimi, M.A, Cusack, S. | | 登録日 | 2015-11-24 | | 公開日 | 2016-08-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Cryptosporidium and Toxoplasma Parasites are Inhibited by a Benzoxaborole Targeting Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
8I4Z
 
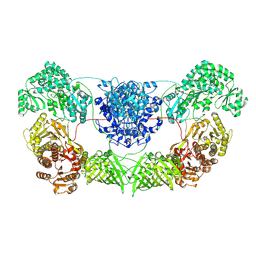 | | CalA3 with hydrolysis product | | 分子名称: | 11-oxidanylidene-11-(1~{H}-pyrrol-2-yl)undecanoic acid, Beta-ketoacyl-acyl-carrier-protein synthase I | | 著者 | Wang, J, Wang, Z. | | 登録日 | 2023-01-21 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.97 Å) | | 主引用文献 | C-N bond formation by a polyketide synthase.
Nat Commun, 14, 2023
|
|
8I4Y
 
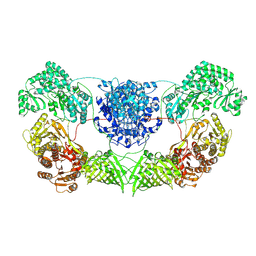 | | CalA3 complex structure with amidation product | | 分子名称: | 11-oxidanylidene-11-(1~{H}-pyrrol-2-yl)undecanoic acid, 3-HYDROXYANTHRANILIC ACID, Beta-ketoacyl-acyl-carrier-protein synthase I | | 著者 | Wang, J, Wang, Z. | | 登録日 | 2023-01-21 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.84 Å) | | 主引用文献 | C-N bond formation by a polyketide synthase.
Nat Commun, 14, 2023
|
|
5FGH
 
 | |
5FON
 
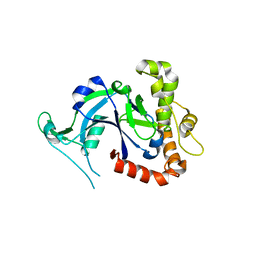 | | Crystal structure of the Cryptosporidium muris cytosolic leucyl-tRNA synthetase editing domain (apo structure) | | 分子名称: | LEUCYL-TRNA SYNTHETASE | | 著者 | Palencia, A, Liu, R.J, Lukarska, M, Gut, J, Bougdour, A, Touquet, B, Wang, E.D, Alley, M.R.K, Rosenthal, P.J, Hakimi, M.A, Cusack, S. | | 登録日 | 2015-11-24 | | 公開日 | 2016-08-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Cryptosporidium and Toxoplasma Parasites are Inhibited by a Benzoxaborole Targeting Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
8FRE
 
 | | Designed loop protein RBL4 | | 分子名称: | 1,2-ETHANEDIOL, RBL4 | | 著者 | Jude, K.M, Jiang, H, Baker, D, Garcia, K.C. | | 登録日 | 2023-01-06 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | De novo design of buttressed loops for sculpting protein functions.
Nat.Chem.Biol., 2024
|
|
8FRF
 
 | |
8FG6
 
 | |
5FGG
 
 | |
