4O26
 
 | |
2G6U
 
 | |
1V6M
 
 | | Peanut Lectin with 9mer peptide (IWSSAGNVA) | | 分子名称: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION | | 著者 | Kundhavai Natchiar, S, Arockia Jeyaprakash, A, Ramya, T.N.C, Thomas, C.J, Suguna, K, Surolia, A, Vijayan, M. | | 登録日 | 2003-12-02 | | 公開日 | 2004-02-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural plasticity of peanut lectin: an X-ray analysis involving variation in pH, ligand binding and crystal structure.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1V6O
 
 | | Peanut lectin complexed with 10mer peptide (PVRIWSSATG) | | 分子名称: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION | | 著者 | Kundhavai Natchiar, S, Arockia Jeyaprakash, A, Ramya, T.N.C, Thomas, C.J, Suguna, K, Surolia, A, Vijayan, M. | | 登録日 | 2003-12-02 | | 公開日 | 2004-02-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural plasticity of peanut lectin: an X-ray analysis involving variation in pH, ligand binding and crystal structure.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OSC
 
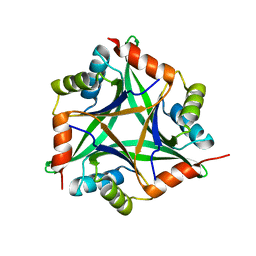 | | Crystal structure of rat CUTA1 at 2.15 A resolution | | 分子名称: | similar to divalent cation tolerant protein CUTA | | 著者 | Arnesano, F, Banci, L, Benvenuti, M, Bertini, I, Calderone, V, Mangani, S, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | 登録日 | 2003-03-19 | | 公開日 | 2003-11-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The Evolutionarily Conserved Trimeric Structure of CutA1 Proteins
Suggests a Role in Signal Transduction
J.Biol.Chem., 278, 2003
|
|
1Q5X
 
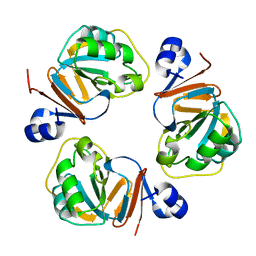 | | Structure of OF RRAA (MENG), a protein inhibitor of RNA processing | | 分子名称: | REGULATOR OF RNASE E ACTIVITY A | | 著者 | Monzingo, A.F, Gao, J, Qiu, J, Georgiou, G, Robertus, J.D. | | 登録日 | 2003-08-11 | | 公開日 | 2003-09-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The X-ray Structure of Escherichia coli RraA (MenG), A Protein Inhibitor of RNA Processing.
J.Mol.Biol., 332, 2003
|
|
4NAA
 
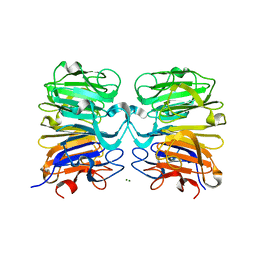 | | Crystal structure of UVB photoreceptor UVR8 from Arabidopsis thaliana and UV-induced structural changes at 120K | | 分子名称: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | 著者 | Yang, X, Zeng, X, Ren, Z, Zhao, K.H. | | 登録日 | 2013-10-22 | | 公開日 | 2016-10-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
1XWI
 
 | |
1V6N
 
 | | Peanut lectin with 9mer peptide (PVIWSSATG) | | 分子名称: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION | | 著者 | Kundhavai Natchiar, S, Arockia Jeyaprakash, A, Ramya, T.N.C, Thomas, C.J, Suguna, K, Surolia, A, Vijayan, M. | | 登録日 | 2003-12-02 | | 公開日 | 2004-02-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural plasticity of peanut lectin: an X-ray analysis involving variation in pH, ligand binding and crystal structure.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4C11
 
 | |
2HE9
 
 | | Structure of the peptidylprolyl isomerase domain of the human NK-tumour recognition protein | | 分子名称: | NK-tumor recognition protein, SULFATE ION | | 著者 | Walker, J.R, Davis, T, Newman, E.M, MacKenzie, F, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | 登録日 | 2006-06-21 | | 公開日 | 2006-07-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases.
PLoS Biol., 8, 2010
|
|
2L1B
 
 | | Solution NMR structure of the chromobox protein Cbx7 with H3K27me3 | | 分子名称: | Chromobox protein homolog 7, Histone H3 | | 著者 | Kaustov, L, Lemak, A, Fares, C, Gutmanas, A, Muhandiram, R, Quang, H, Loppnau, P, Min, J, Edwards, A, Arrowsmith, C, Structural Genomics Consortium (SGC) | | 登録日 | 2010-07-27 | | 公開日 | 2010-08-25 | | 最終更新日 | 2020-02-05 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
4K2M
 
 | | Crystal structure of ntda from bacillus subtilis in complex with the plp external aldimine adduct with kanosamine-6-phosphate | | 分子名称: | 1,2-ETHANEDIOL, 3-deoxy-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-O-phosphono-alpha-D-gluco pyranose, ACETATE ION, ... | | 著者 | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | 登録日 | 2013-04-09 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | The Structure of NtdA, a Sugar Aminotransferase Involved in the Kanosamine Biosynthetic Pathway in Bacillus subtilis, Reveals a New Subclass of Aminotransferases.
J.Biol.Chem., 288, 2013
|
|
4K2B
 
 | |
4KBK
 
 | |
4K2I
 
 | | Crystal structure of ntda from bacillus subtilis with bound cofactor pmp | | 分子名称: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ACETATE ION, ... | | 著者 | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | 登録日 | 2013-04-09 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.225 Å) | | 主引用文献 | The Structure of NtdA, a Sugar Aminotransferase Involved in the Kanosamine Biosynthetic Pathway in Bacillus subtilis, Reveals a New Subclass of Aminotransferases.
J.Biol.Chem., 288, 2013
|
|
4KBC
 
 | |
4KB8
 
 | |
4KZ6
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 13 ((2R,6R)-6-methyl-1-(3-sulfanylpropanoyl)piperidine-2-carboxylic acid) | | 分子名称: | (2R,6R)-6-methyl-1-(3-sulfanylpropanoyl)piperidine-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | 著者 | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | 登録日 | 2013-05-29 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZA
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 48 (3-(cyclopropylsulfamoyl)thiophene-2-carboxylic acid) | | 分子名称: | 3-(cyclopropylsulfamoyl)thiophene-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | 著者 | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | 登録日 | 2013-05-29 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZ7
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 16 ((1R,4S)-4,7,7-trimethyl-3-oxo-2-oxabicyclo[2.2.1]heptane-1-carboxylic acid) | | 分子名称: | (1R,4S)-4,7,7-trimethyl-3-oxo-2-oxabicyclo[2.2.1]heptane-1-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | 著者 | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | 登録日 | 2013-05-29 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZ3
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 44 (5-chloro-3-sulfamoylthiophene-2-carboxylic acid) | | 分子名称: | 5-chloro-3-sulfamoylthiophene-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | 著者 | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | 登録日 | 2013-05-29 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZB
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 50 (N-(methylsulfonyl)-N-phenyl-alanine) | | 分子名称: | Beta-lactamase, N-(methylsulfonyl)-N-phenyl-D-alanine, N-(methylsulfonyl)-N-phenyl-L-alanine | | 著者 | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | 登録日 | 2013-05-29 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZ8
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 20 (1,3-diethyl-2-thioxodihydropyrimidine-4,6(1H,5H)-dione) | | 分子名称: | 1,3-diethyl-2-thioxodihydropyrimidine-4,6(1H,5H)-dione, Beta-lactamase, PHOSPHATE ION | | 著者 | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | 登録日 | 2013-05-29 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.282 Å) | | 主引用文献 | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZ4
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 60 (2-[(propylsulfonyl)amino]benzoic acid) | | 分子名称: | 2-[(propylsulfonyl)amino]benzoic acid, Beta-lactamase, PHOSPHATE ION | | 著者 | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | 登録日 | 2013-05-29 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
