1L4Y
 
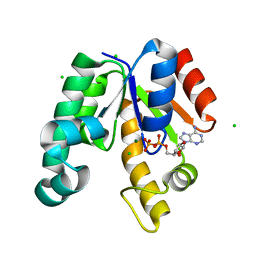 | | CRYSTAL STRUCTURE OF SHIKIMATE KINASE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MGADP AT 2.0 ANGSTROM RESOLUTION | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Gu, Y, Reshetnikova, L, Li, Y, Wu, Y, Yan, H, Singh, S, Ji, X. | | 登録日 | 2002-03-06 | | 公開日 | 2002-06-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of shikimate kinase from Mycobacterium tuberculosis reveals the dynamic role of the LID domain in catalysis.
J.Mol.Biol., 319, 2002
|
|
1LB6
 
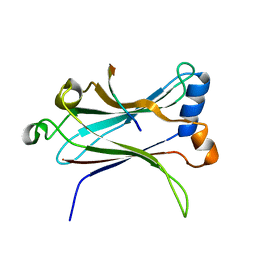 | | TRAF6-CD40 Complex | | 分子名称: | CD40 antigen, TNF receptor-associated factor 6 | | 著者 | Ye, H, Arron, J.R, Lamothe, B, Cirilli, M, Kobayashi, T, Shevde, N.K, Segal, D, Dzivenu, O, Vologodskaia, M, Yim, M, Du, K, Singh, S, Pike, J.W, Darnay, B.G, Choi, Y, Wu, H. | | 登録日 | 2002-04-02 | | 公開日 | 2002-07-31 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Distinct molecular mechanism for initiating TRAF6 signalling.
Nature, 418, 2002
|
|
1M7Q
 
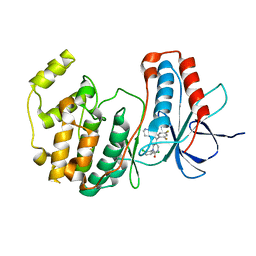 | | Crystal structure of p38 MAP kinase in complex with a dihydroquinazolinone inhibitor | | 分子名称: | 1-(2,6-DICHLOROPHENYL)-5-(2,4-DIFLUOROPHENYL)-7-PIPERAZIN-1-YL-3,4-DIHYDROQUINAZOLIN-2(1H)-ONE, Mitogen-activated protein kinase 14, SULFATE ION | | 著者 | Stelmach, J.E, Liu, L, Patel, S.B, Pivnichny, J.V, Scapin, G, Singh, S, Hop, C.E.C.A, Wang, Z, Cameron, P.M, Nichols, E.A, O'Keefe, S.J, O'Neill, E.A, Schmatz, D.M, Schwartz, C.D, Thompson, C.M, Zaller, D.M, Doherty, J.B. | | 登録日 | 2002-07-22 | | 公開日 | 2002-12-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Design and synthesis of potent, orally bioavailable dihydroquinazolinone inhibitors of p38 MAP kinase.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
4L7I
 
 | | Crystal structure of S-Adenosylmethionine synthase from Sulfolobus solfataricus complexed with SAM and PPi | | 分子名称: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2013-06-13 | | 公開日 | 2013-07-03 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.189 Å) | | 主引用文献 | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
4P8E
 
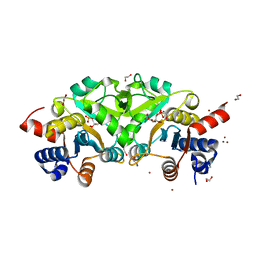 | | Structure of ribB complexed with substrate (Ru5P) and metal ions | | 分子名称: | 1,2-ETHANEDIOL, 3,4-dihydroxy-2-butanone 4-phosphate synthase, RIBULOSE-5-PHOSPHATE, ... | | 著者 | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | 登録日 | 2014-03-31 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
4P6C
 
 | | Structure of ribB complexed with inhibitor 4PEH | | 分子名称: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, 4-PHOSPHO-D-ERYTHRONOHYDROXAMIC ACID | | 著者 | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | 登録日 | 2014-03-24 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
3OTG
 
 | | Crystal Structure of CalG1, Calicheamicin Glycostyltransferase, TDP bound form | | 分子名称: | CHLORIDE ION, CalG1, THYMIDINE-5'-DIPHOSPHATE | | 著者 | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2010-09-11 | | 公開日 | 2010-12-15 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PAU
 
 | |
3OTI
 
 | | Crystal Structure of CalG3, Calicheamicin Glycostyltransferase, TDP and calicheamicin T0 bound form | | 分子名称: | CHLORIDE ION, CalG3, Calicheamicin T0, ... | | 著者 | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2010-09-11 | | 公開日 | 2010-12-15 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.597 Å) | | 主引用文献 | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OTH
 
 | | Crystal Structure of CalG1, Calicheamicin Glycostyltransferase, TDP and calicheamicin alpha3I bound form | | 分子名称: | CalG1, Calicheamicin alpha3I, THYMIDINE-5'-DIPHOSPHATE | | 著者 | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2010-09-11 | | 公開日 | 2010-12-15 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4M83
 
 | | Ensemble refinement of protein crystal structure (2IYF) of macrolide glycosyltransferases OleD complexed with UDP and Erythromycin A | | 分子名称: | ERYTHROMYCIN A, MAGNESIUM ION, Oleandomycin glycosyltransferase, ... | | 著者 | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2013-08-12 | | 公開日 | 2013-09-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.698 Å) | | 主引用文献 | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
2OA1
 
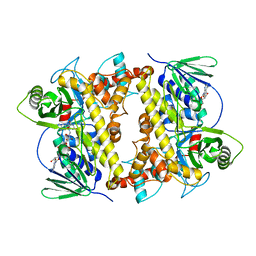 | | Crystal Structure of RebH, a FAD-dependent halogenase from Lechevalieria aerocolonigenes, the L-Tryptophan with FAD complex | | 分子名称: | ADENOSINE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Bitto, E, Bingman, C.A, Singh, S, Phillips Jr, G.N. | | 登録日 | 2006-12-14 | | 公開日 | 2007-04-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The structure of flavin-dependent tryptophan 7-halogenase RebH.
Proteins, 70, 2008
|
|
2EVN
 
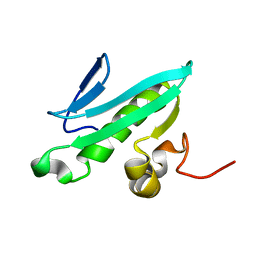 | | NMR solution structures of At1g77540 | | 分子名称: | Protein At1g77540 | | 著者 | Tyler, R.C, Singh, S, Tonelli, M, Min, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2005-10-31 | | 公開日 | 2005-11-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
4FZR
 
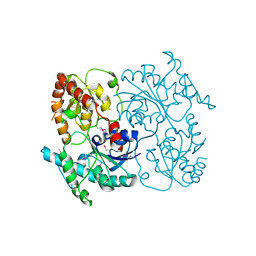 | | Crystal Structure of SsfS6, Streptomyces sp. SF2575 glycosyltransferase | | 分子名称: | SsfS6 | | 著者 | Wang, F, Zhou, M, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2012-07-07 | | 公開日 | 2012-07-25 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.397 Å) | | 主引用文献 | Crystal structure of SsfS6, the putative C-glycosyltransferase involved in SF2575 biosynthesis.
Proteins, 81, 2013
|
|
4G2T
 
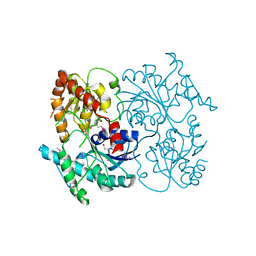 | | Crystal Structure of Streptomyces sp. SF2575 glycosyltransferase SsfS6, complexed with thymidine diphosphate | | 分子名称: | SsfS6, THYMIDINE-5'-DIPHOSPHATE | | 著者 | Wang, F, Zhou, M, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2012-07-12 | | 公開日 | 2012-07-25 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.405 Å) | | 主引用文献 | Crystal structure of SsfS6, the putative C-glycosyltransferase involved in SF2575 biosynthesis.
Proteins, 81, 2013
|
|
3T6U
 
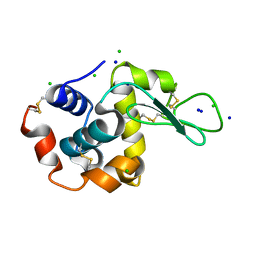 | |
3RSC
 
 | | Crystal Structure of CalG2, Calicheamicin Glycosyltransferase, TDP and calicheamicin T0 bound form | | 分子名称: | CalG2, Calicheamicin T0, PHOSPHATE ION, ... | | 著者 | Chang, A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2011-05-02 | | 公開日 | 2011-08-10 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4M60
 
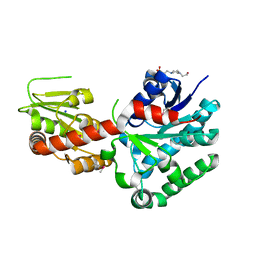 | | Crystal structure of macrolide glycosyltransferases OleD | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Oleandomycin glycosyltransferase, SODIUM ION | | 著者 | Olmos Jr, J.L, Martinez III, E, Wang, F, Helmich, K.E, Singh, S, Xu, W, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2013-08-08 | | 公開日 | 2013-09-04 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
4L2Z
 
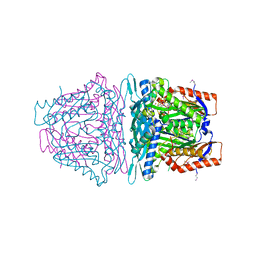 | | Crystal structure of S-Adenosylmethionine synthetase from Sulfolobus solfataricus complexed with SAE and PPi | | 分子名称: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2013-06-05 | | 公開日 | 2013-06-19 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.494 Å) | | 主引用文献 | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
4M7P
 
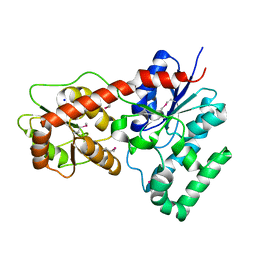 | | Ensemble refinement of protein crystal structure of macrolide glycosyltransferases OleD | | 分子名称: | Oleandomycin glycosyltransferase, SODIUM ION | | 著者 | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2013-08-12 | | 公開日 | 2013-09-11 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
5ZZ0
 
 | | HUMAN GELSOLIN FROM RESIDUES GLU28 TO ARG161 WITH CALCIUM | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, Gelsolin | | 著者 | Sharma, P, Badmalia, M, Yadav, S.P.S, Singh, S. | | 登録日 | 2018-05-29 | | 公開日 | 2019-06-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.635 Å) | | 主引用文献 | Bonsai Gelsolin Survives Heat Induced Denaturation by Forming beta-Amyloids which Leach Out Functional Monomer.
Sci Rep, 8, 2018
|
|
4PIW
 
 | | Crystal structure of sugar aminotransferase WecE from Escherichia coli K-12 | | 分子名称: | TDP-4-keto-6-deoxy-D-glucose transaminase family protein | | 著者 | Wang, F, Xu, W, Helmich, K.E, Singh, S, Yennamalli, R.M, Miller, M.D, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2014-05-09 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of sugar aminotransferase WecE from Escherichia coli K-12
To Be Published
|
|
2IVW
 
 | | The solution structure of a domain from the Neisseria meningitidis PilP pilot protein. | | 分子名称: | PILP PILOT PROTEIN | | 著者 | Golovanov, A.P, Balasingham, S, Tzitzilonis, C, Goult, B.T, Lian, L.-Y, Homberset, H, Tonjum, T, Derrick, J.P. | | 登録日 | 2006-06-20 | | 公開日 | 2007-02-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of a domain from the Neisseria meningitidis lipoprotein PilP reveals a new beta-sandwich fold.
J. Mol. Biol., 364, 2006
|
|
4ZWV
 
 | | Crystal Structure of Aminotransferase AtmS13 from Actinomadura melliaura | | 分子名称: | GLYCEROL, Putative aminotransferase | | 著者 | Kim, Y, Bigelow, L, Endres, M, Wang, F, Phillips Jr, G.N, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-05-19 | | 公開日 | 2015-06-03 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (1.503 Å) | | 主引用文献 | Structural characterization of AtmS13, a putative sugar aminotransferase involved in indolocarbazole AT2433 aminopentose biosynthesis.
Proteins, 83, 2015
|
|
1NPY
 
 | | Structure of shikimate 5-dehydrogenase-like protein HI0607 | | 分子名称: | ACETYL GROUP, Hypothetical shikimate 5-dehydrogenase-like protein HI0607 | | 著者 | Korolev, S, Koroleva, O, Zarembinski, T, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2003-01-20 | | 公開日 | 2003-07-29 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal Structure of a Novel Shikimate Dehydrogenase from Haemophilus influenzae.
J.Biol.Chem., 280, 2005
|
|
