5SUZ
 
 | |
1UU4
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMICOLA GRISEA CEL12A IN COMPLEX WITH CELLOBIOSE | | 分子名称: | ENDO-BETA-1,4-GLUCANASE, TETRAETHYLENE GLYCOL, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Berglund, G.I, Shaw, A, Stahlberg, J, Kenne, L, Driguez, T.H, Mitchinson, C, Sandgren, M. | | 登録日 | 2003-12-15 | | 公開日 | 2004-09-16 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Crystal Complex Structures Reveal How Substrate is Bound in the -4 to the +2 Binding Sites of Humicola Grisea Cel12A
J.Mol.Biol., 342, 2004
|
|
1UU5
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMICOLA GRISEA CEL12A SOAKED WITH CELLOTETRAOSE | | 分子名称: | ACETATE ION, ENDO-BETA-1,4-GLUCANASE, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Berglund, G.I, Shaw, A, Stahlberg, J, Kenne, L, Driguez, T.H, Mitchinson, C, Sandgren, M. | | 登録日 | 2003-12-15 | | 公開日 | 2004-09-16 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Crystal Complex Structures Reveal How Substrate is Bound in the -4 to the +2 Binding Sites of Humicola Grisea Cel12A
J.Mol.Biol., 342, 2004
|
|
1QUR
 
 | |
9FM9
 
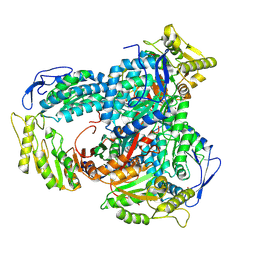 | |
9FLZ
 
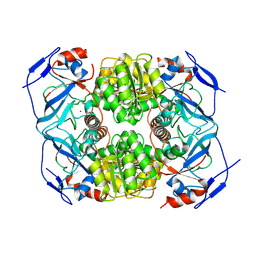 | |
5JU6
 
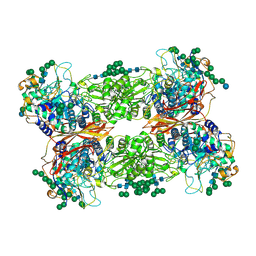 | | Structural and Functional Studies of Glycoside Hydrolase Family 3 beta-Glucosidase Cel3A from the Moderately Thermophilic Fungus Rasamsonia emersonii | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-glucosidase, ... | | 著者 | Gudmundsson, M, Sandgren, M, Karkehabadi, S. | | 登録日 | 2016-05-10 | | 公開日 | 2016-07-13 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and functional studies of the glycoside hydrolase family 3 beta-glucosidase Cel3A from the moderately thermophilic fungus Rasamsonia emersonii.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4CYE
 
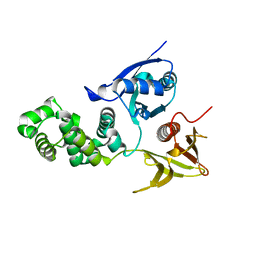 | | Crystal structure of avian FAK FERM domain FAK31-405 at 3.2A | | 分子名称: | FOCAL ADHESION KINASE 1 | | 著者 | Goni, G.M, Epifano, C, Boskovic, J, Camacho-Artacho, M, Zhou, J, Martin, M.T, Eck, M.J, Kremer, L, Graeter, F, Gervasio, F.L, Perez-Moreno, M, Lietha, D. | | 登録日 | 2014-04-10 | | 公開日 | 2014-04-23 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Phosphatidylinositol 4,5-Bisphosphate Triggers Activation of Focal Adhesion Kinase by Inducing Clustering and Conformational Changes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8RKS
 
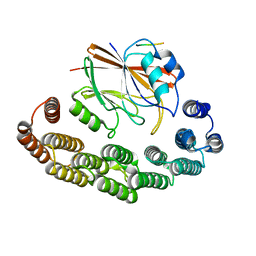 | | Structure of VPS29-VPS35 bound to the LFa motif R21 of Fam21. | | 分子名称: | Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35, WASH complex subunit 2A | | 著者 | Romano-Moreno, M, Astorga-Simon, E.N, Rojas, A.L, Hierro, A. | | 登録日 | 2023-12-30 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Retromer-mediated recruitment of the WASH complex involves discrete interactions between VPS35, VPS29, and FAM21.
Protein Sci., 33, 2024
|
|
4D7U
 
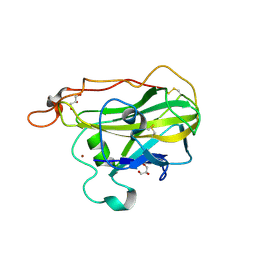 | | The structure of the catalytic domain of NcLPMO9C from the filamentous fungus Neurospora crassa | | 分子名称: | COPPER (II) ION, ENDOGLUCANASE II, GLYCEROL | | 著者 | Borisova, A.S, Isaksen, T, Mathiesen, G, Sorlie, M, Sandgren, M, Eijsink, V.G.H, Dimarogona, M. | | 登録日 | 2014-11-27 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Structural and Functional Characterization of a Lytic Polysaccharide Monooxygenase with Broad Substrate Specificity
J.Biol.Chem., 290, 2015
|
|
4D7V
 
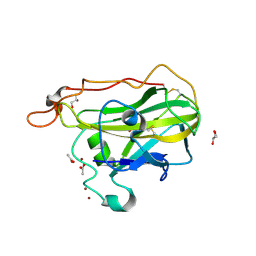 | | The structure of the catalytic domain of NcLPMO9C from the filamentous fungus Neurospora crassa | | 分子名称: | ACETATE ION, ENDOGLUCANASE II, GLYCEROL, ... | | 著者 | Borisova, A.S, Isaksen, T, Sandgren, M, Sorlie, M, Eijsink, V.G.H, Dimarogona, M. | | 登録日 | 2014-11-27 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and Functional Characterization of a Lytic Polysaccharide Monooxygenase with Broad Substrate Specificity
J.Biol.Chem., 290, 2015
|
|
6T8U
 
 | | Complement factor B in complex with 5-Bromo-3-chloro-N-(4,5-dihydro-1H-imidazol-2-yl)-7-methyl-1H-indol-4-amine | | 分子名称: | 5-bromanyl-3-chloranyl-~{N}-(1~{H}-imidazol-2-yl)-7-methyl-1~{H}-indol-4-amine, Complement factor B, SULFATE ION | | 著者 | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | 登録日 | 2019-10-25 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
4V69
 
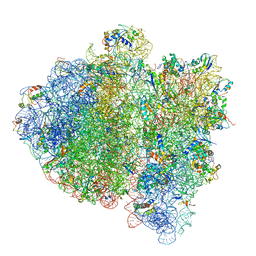 | | Ternary complex-bound E.coli 70S ribosome. | | 分子名称: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Villa, E, Sengupta, J, Trabuco, L.G, LeBarron, J, Baxter, W.T, Shaikh, T.R, Grassucci, R.A, Nissen, P, Ehrenberg, M, Schulten, K, Frank, J. | | 登録日 | 2008-12-11 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (6.7 Å) | | 主引用文献 | Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6T8V
 
 | | Complement factor B in complex with (S)-5,7-Dimethyl-4-((2-phenylpiperidin-1-yl)methyl)-1H-indole | | 分子名称: | 4-[(2~{S})-1-[(5,7-dimethyl-1~{H}-indol-4-yl)methyl]piperidin-2-yl]benzoic acid, Complement factor B, SULFATE ION, ... | | 著者 | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | 登録日 | 2019-10-25 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
6EXM
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (MK202) | | 分子名称: | ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, SODIUM ION, ... | | 著者 | Grundstrom, C, Begum, A, Hall, M, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | 登録日 | 2017-11-08 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
6EUT
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (KSK67) | | 分子名称: | (3~{R})-8-cyclopropyl-7-[(4-methylnaphthalen-1-yl)methyl]-5-oxidanylidene-2,3-dihydro-[1,3]thiazolo[3,2-a]pyridine-3-carboxylic acid, GLYCEROL, ISOPROPYL ALCOHOL, ... | | 著者 | Begum, A, Hall, M, Grundstrom, C, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | 登録日 | 2017-10-31 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
6EUZ
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (MK37) | | 分子名称: | (3~{R})-8-methoxy-7-(naphthalen-1-ylmethyl)-5-oxidanylidene-2,3-dihydro-[1,3]thiazolo[3,2-a]pyridine-3-carboxylic acid, ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, ... | | 著者 | Begum, A, Hall, M, Grundstrom, C, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | 登録日 | 2017-10-31 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
6EXL
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (MK206) - folded HTH motif | | 分子名称: | ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, SODIUM ION, ... | | 著者 | Hall, M, Grundstrom, C, Begum, A, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | 登録日 | 2017-11-08 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
6EUU
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (KSK29) | | 分子名称: | (3~{R})-8-cyclopropyl-5-oxidanylidene-7-(quinolin-3-ylmethyl)-2,3-dihydro-[1,3]thiazolo[3,2-a]pyridine-3-carboxylic acid, ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, ... | | 著者 | Begum, A, Hall, M, Grundstrom, C, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | 登録日 | 2017-10-31 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
6EV0
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (AC129) | | 分子名称: | ISOPROPYL ALCOHOL, Listeriolysin regulatory protein, [(3~{R})-3-carboxy-7-(naphthalen-1-ylmethyl)-5-oxidanylidene-2,3-dihydro-[1,3]thiazolo[3,2-a]pyridin-8-yl]-dimethyl-azanium, ... | | 著者 | Begum, A, Hall, M, Grundstrom, C, Cairns, A.G, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | 登録日 | 2017-10-31 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
6EXK
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (MK206) - unfolded HTH motif | | 分子名称: | ISOPROPYL ALCOHOL, Listeriolysin regulatory protein, SODIUM ION, ... | | 著者 | Hall, M, Grundstrom, C, Begum, A, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | 登録日 | 2017-11-08 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
8RZL
 
 | | Sulfolobus acidocaldarius threads (0406) filament. | | 分子名称: | Sulfolobus acidocaldarius threads (0406) filament., alpha-D-mannopyranose, beta-D-glucopyranose-(1-4)-6-deoxy-6-sulfo-beta-D-glucopyranose-(1-3)-[alpha-D-mannopyranose-(1-4)][alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Isupov, M.N, Gaines, M, McLaren, M, Daum, B. | | 登録日 | 2024-02-12 | | 公開日 | 2024-05-01 | | 最終更新日 | 2025-02-05 | | 実験手法 | ELECTRON MICROSCOPY (2.65 Å) | | 主引用文献 | Towards a molecular picture of the archaeal cell surface.
Nat Commun, 15, 2024
|
|
4CSA
 
 | | Crystal structure of the asymmetric human metapneumovirus M2-1 tetramer bound to a DNA 4-mer | | 分子名称: | 5'-D(*AP*GP*TP*TP*AP)-3', GLYCEROL, M2-1, ... | | 著者 | Leyrat, C, Renner, M, Harlos, K, Grimes, J.M. | | 登録日 | 2014-03-05 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Drastic Changes in Conformational Dynamics of the Antiterminator M2-1 Regulate Transcription Efficiency in Pneumovirinae.
Elife, 3, 2014
|
|
8GY6
 
 | |
9QK3
 
 | | X-ray crystal structure of SlPYL1-Coumaric Acid complex | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, DIMETHYL SULFOXIDE, SlPYL1-NIO | | 著者 | Rivera-Moreno, M, Merino-Gracia, J, Infantes, L, Albert, A. | | 登録日 | 2025-03-19 | | 公開日 | 2025-07-02 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Natural modulators of abscisic acid Signaling: Insights into polyphenol-based antagonists and their role in ABA receptor regulation.
Plant Physiol Biochem., 227, 2025
|
|
