6ECN
 
 | | HIV-1 CA 1/2-hexamer-EE | | 分子名称: | HIV-1 CA | | 著者 | Summers, B.J, Xiong, Y. | | 登録日 | 2018-08-08 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
6EC2
 
 | | Structure of HIV-1 CA 1/3-hexamer | | 分子名称: | ACETATE ION, Capsid protein p24 | | 著者 | Summers, B.J, Xiong, Y. | | 登録日 | 2018-08-07 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
6ECO
 
 | |
5LZN
 
 | | -TIP microtubule-binding domain | | 分子名称: | Calmodulin-regulated spectrin-associated protein 3 | | 著者 | Stangier, M.M, Steinmetz, M.O. | | 登録日 | 2016-09-30 | | 公開日 | 2017-10-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | A structural model for microtubule minus-end recognition and protection by CAMSAP proteins.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6TY9
 
 | | In situ structure of BmCPV RNA dependent RNA polymerase at initiation state | | 分子名称: | MAGNESIUM ION, Non-template RNA (5'-D(*(GTA))-R(P*GP*UP*AP*AP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), RNA-dependent RNA Polymerase, ... | | 著者 | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | 登録日 | 2019-08-08 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6TZ1
 
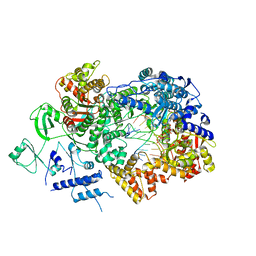 | | In situ structure of BmCPV RNA-dependent RNA polymerase at early-elongation state | | 分子名称: | Non-template RNA (5'-D(*(GTA))-R(P*GP*UP*A)-3'), RNA-dependent RNA Polymerase, Template RNA (5'-R(P*AP*GP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | 著者 | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | 登録日 | 2019-08-09 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6TY8
 
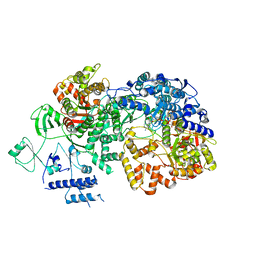 | | In situ structure of BmCPV RNA dependent RNA polymerase at quiescent state | | 分子名称: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA-dependent RNA Polymerase, Viral structural protein 4 | | 著者 | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | 登録日 | 2019-08-08 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6TZ2
 
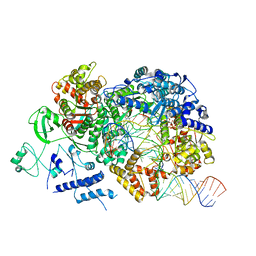 | | In situ structure of BmCPV RNA-dependent RNA polymerase at elongation state | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Non-template RNA (36-MER), ... | | 著者 | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | 登録日 | 2019-08-09 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6TZ0
 
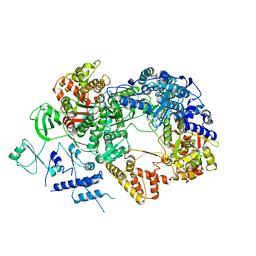 | | In situ structure of BmCPV RNA-dependent RNA polymerase at abortive state | | 分子名称: | RNA-dependent RNA Polymerase, Viral structural protein 4 | | 著者 | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | 登録日 | 2019-08-09 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6UX2
 
 | |
4LMA
 
 | |
4LMB
 
 | |
3O05
 
 | | Crystal Structure of Yeast Pyridoxal 5-Phosphate Synthase Snz1 Complxed with Substrate PLP | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxine biosynthesis protein SNZ1 | | 著者 | Teng, Y.B, Zhang, X, He, Y.X, Hu, H.X, Zhou, C.Z. | | 登録日 | 2010-07-19 | | 公開日 | 2010-11-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural insights into the catalytic mechanism of the yeast pyridoxal 5-phosphate synthase Snz1
Biochem.J., 432, 2010
|
|
3O06
 
 | |
3O07
 
 | | Crystal structure of yeast pyridoxal 5-phosphate synthase Snz1 complexed with substrate G3P | | 分子名称: | GLYCERALDEHYDE-3-PHOSPHATE, Pyridoxine biosynthesis protein SNZ1 | | 著者 | Teng, Y.B, Zhang, X, Hu, H.X, Zhou, C.Z. | | 登録日 | 2010-07-19 | | 公開日 | 2010-11-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into the catalytic mechanism of the yeast pyridoxal 5-phosphate synthase Snz1
Biochem.J., 432, 2010
|
|
8H3Z
 
 | |
8H3V
 
 | | Cryo-EM structure of the full transcription activation complex NtcA-NtcB-TAC | | 分子名称: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | 登録日 | 2022-10-09 | | 公開日 | 2023-10-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H40
 
 | | Cryo-EM structure of the transcription activation complex NtcA-TAC | | 分子名称: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | 登録日 | 2022-10-09 | | 公開日 | 2023-10-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6UFR
 
 | | Structure of recombinantly assembled E46K alpha-synuclein fibrils | | 分子名称: | Alpha-synuclein | | 著者 | Eisenberg, D.S, Boyer, D.R, Sawaya, M.R, Li, B, Jiang, L. | | 登録日 | 2019-09-24 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | The alpha-synuclein hereditary mutation E46K unlocks a more stable, pathogenic fibril structure.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8GN6
 
 | |
6KXG
 
 | |
4MDN
 
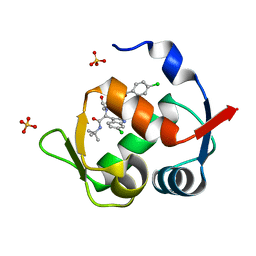 | | Structure of a novel submicromolar MDM2 inhibitor | | 分子名称: | 3-{(1S)-2-(tert-butylamino)-1-[{4-[(4-chlorobenzyl)oxy]benzyl}(formyl)amino]-2-oxoethyl}-6-chloro-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | 著者 | Bista, M, Popowicz, G, Holak, T.A. | | 登録日 | 2013-08-23 | | 公開日 | 2013-11-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.905 Å) | | 主引用文献 | Transient Protein States in Designing Inhibitors of the MDM2-p53 Interaction.
Structure, 21, 2013
|
|
5F9R
 
 | |
4MDQ
 
 | | Structure of a novel submicromolar MDM2 inhibitor | | 分子名称: | 3-[(1R)-2-(benzylamino)-1-{[(2S)-1-(hydroxyamino)-4-methyl-1-oxopentan-2-yl]amino}-2-oxoethyl]-6-chloro-N-hydroxy-1H-indole-2-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | 著者 | Bista, M, Popowicz, G, Holak, T.A. | | 登録日 | 2013-08-23 | | 公開日 | 2013-11-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.119 Å) | | 主引用文献 | Transient Protein States in Designing Inhibitors of the MDM2-p53 Interaction.
Structure, 21, 2013
|
|
5XNJ
 
 | |
