1VDD
 
 | |
1R5H
 
 | | Crystal Structure of MetAP2 complexed with A320282 | | 分子名称: | MANGANESE (II) ION, Methionine aminopeptidase 2, N'-(2S,3R)-3-AMINO-4-CYCLOHEXYL-2-HYDROXY-BUTANO-N-(4-METHYLPHENYL)HYDRAZIDE | | 著者 | Sheppard, G.S, Wang, J, Kawai, M, BaMaung, N.Y, Craig, R.A, Erickson, S.A, Lynch, L, Patel, J, Yang, F, Searle, X.B, Lou, P, Park, C, Kim, K.H, Henkin, J, Lesniewski, R. | | 登録日 | 2003-10-10 | | 公開日 | 2004-10-12 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1R58
 
 | | Crystal Structure of MetAP2 complexed with A357300 | | 分子名称: | MANGANESE (II) ION, Methionine aminopeptidase 2, N'-((2S,3R)-3-AMINO-2-HYDROXY-5-(ISOPROPYLSULFANYL)PENTANOYL)-N-3-CHLOROBENZOYL HYDRAZIDE | | 著者 | Sheppard, G.S, Wang, J, Kawai, M, BaMaung, N.Y, Craig, R.A, Ericken, S.A, Lynch, L, Patel, J, Yang, F, Searle, X.B, Lou, P, Park, C, Kim, K.H, Henkin, J, Lesniewski, R. | | 登録日 | 2003-10-09 | | 公開日 | 2004-10-12 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
6KYV
 
 | | Crystal Structure of RIG-I and hairpin RNA with G-U wobble base pairs | | 分子名称: | Probable ATP-dependent RNA helicase DDX58, RNA (5'-R(*GP*GP*UP*AP*GP*AP*CP*GP*CP*UP*UP*CP*GP*GP*CP*GP*UP*UP*UP*GP*CP*C)-3'), ZINC ION | | 著者 | Kim, K.-H, Hwang, J, Kim, J.H, Son, K.-P, Jang, Y, Kim, M, Kang, S.-J, Lee, J.-O, Choi, B.-S. | | 登録日 | 2019-09-20 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural and biophysical properties of RIG-I bound to dsRNA with G-U wobble base pairs.
Rna Biol., 17, 2020
|
|
5HKN
 
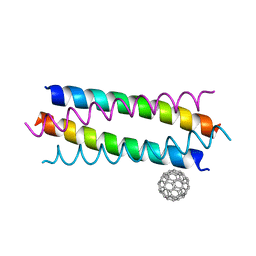 | | Crystal structure de novo designed fullerene organizing protein complex with fullerene | | 分子名称: | (C_{60}-I_{h})[5,6]fullerene, fullerene organizing protein | | 著者 | Paul, J, Acharya, R, Kim, K.-H, Kim, Y.H, Kim, N.H, Grigoryan, G, DeGardo, W.F. | | 登録日 | 2016-01-14 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.761 Å) | | 主引用文献 | Protein-directed self-assembly of a fullerene crystal.
Nat Commun, 7, 2016
|
|
5HKR
 
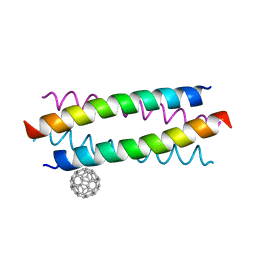 | | Crystal structure of de novo designed fullerene organising protein complex with fullerene | | 分子名称: | (C_{60}-I_{h})[5,6]fullerene, fullerene organizing protein | | 著者 | Acharya, R, Kim, Y.H, Grigoryan, G, DeGardo, W.F. | | 登録日 | 2016-01-14 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Protein-directed self-assembly of a fullerene crystal.
Nat Commun, 7, 2016
|
|
3RBY
 
 | |
2OHV
 
 | |
2OHO
 
 | |
2OHG
 
 | |
2KXX
 
 | | NMR Structure of Escherichia coli BamE, a Lipoprotein Component of the beta-Barrel Assembly Machinery Complex | | 分子名称: | Small protein A | | 著者 | Kim, K, Okon, M, Escobar, E, Kang, H, McIntosh, L, Paetzel, M. | | 登録日 | 2010-05-13 | | 公開日 | 2011-01-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Characterization of Escherichia coli BamE, a Lipoprotein Component of the beta-Barrel Assembly Machinery Complex.
Biochemistry, 50, 2011
|
|
4PQQ
 
 | | The crystal structure of discoidin domain from muskelin | | 分子名称: | Muskelin, PHOSPHATE ION, TETRAETHYLENE GLYCOL | | 著者 | Kim, K.-H, Hong, S.K, Kim, E.E. | | 登録日 | 2014-03-04 | | 公開日 | 2014-11-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structure of mouse muskelin discoidin domain and biochemical characterization of its self-association.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8HG9
 
 | |
3H9O
 
 | |
1NCO
 
 | | STRUCTURE OF THE ANTITUMOR PROTEIN-CHROMOPHORE COMPLEX NEOCARZINOSTATIN | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, HOLO-NEOCARZINOSTATIN, NEOCARZINOSTATIN-CHROMOPHORE | | 著者 | Kim, K.-H, Kwon, B.-M, Myers, A.G, Rees, D.C. | | 登録日 | 1993-07-13 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of neocarzinostatin, an antitumor protein-chromophore complex.
Science, 262, 1993
|
|
7ENY
 
 | | Crystal structure of hydroxysteroid dehydrogenase from Escherichia coli | | 分子名称: | 7alpha-hydroxysteroid dehydrogenase | | 著者 | Kim, K.-H, Lee, C.W, Pardhe, D.P, Hwang, J, Do, H, Lee, Y.M, Lee, J.H, Oh, T.-J. | | 登録日 | 2021-04-21 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.703 Å) | | 主引用文献 | Crystal structure of an apo 7 alpha-hydroxysteroid dehydrogenase reveals key structural changes induced by substrate and co-factor binding.
J.Steroid Biochem.Mol.Biol., 212, 2021
|
|
3T9T
 
 | | Crystal structure of BTK mutant (F435T,K596R) complexed with Imidazo[1,5-a]quinoxaline | | 分子名称: | (2Z)-4-(dimethylamino)-N-{7-fluoro-4-[(2-methylphenyl)amino]imidazo[1,5-a]quinoxalin-8-yl}-N-methylbut-2-enamide, GLYCEROL, Tyrosine-protein kinase ITK/TSK | | 著者 | Han, S, Caspers, N. | | 登録日 | 2011-08-03 | | 公開日 | 2011-10-12 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Imidazo[1,5-a]quinoxalines as irreversible BTK inhibitors for the treatment of rheumatoid arthritis.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5XNT
 
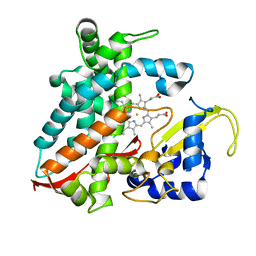 | | Structure of CYP106A2 from Bacillus sp. PAMC 23377 | | 分子名称: | Cytochrome P450 CYP106, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Lee, C.W, Kim, K.-H, Bikash, D, Park, S.-H, Park, H, Oh, T.-J, Lee, J.H. | | 登録日 | 2017-05-24 | | 公開日 | 2018-04-04 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal Structure and Functional Characterization of a Cytochrome P450 (BaCYP106A2) fromBacillussp. PAMC 23377.
J. Microbiol. Biotechnol., 27, 2017
|
|
3QCA
 
 | |
2QKY
 
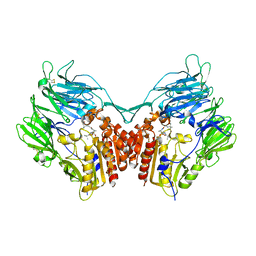 | | complex structure of dipeptidyl peptidase IV and a oxadiazolyl ketone | | 分子名称: | 2-[(2-{(2S,4S)-2-[(R)-(5-tert-butyl-1,3,4-oxadiazol-2-yl)(hydroxy)methyl]-4-fluoropyrrolidin-1-yl}-2-oxoethyl)amino]-2-methylpropan-1-ol, Dipeptidyl peptidase 4 (EC 3.4.14.5) (Dipeptidyl peptidase IV) (DPP IV) (T-cell activation antigen CD26) (TP103) (Adenosine deaminase complexing protein 2) (ADABP) (Dipeptidyl peptidase 4 soluble form) (Dipeptidyl peptidase IV soluble form) | | 著者 | Kim, K.-H, Hong, S.Y, Koo, K.D, Lee, C.-S, Kim, G.T, Han, H.O. | | 登録日 | 2007-07-12 | | 公開日 | 2008-07-15 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Synthesis, SAR, and X-ray structure of novel potent DPPIV inhibitors: oxadiazolyl ketones.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6XAV
 
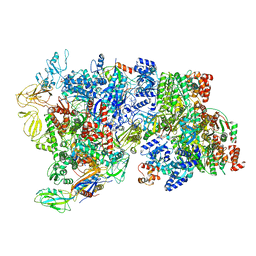 | | CryoEM Structure of E. coli Rho-dependent Transcription Pre-termination Complex bound with NusG | | 分子名称: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Hao, Z.T, Kim, H.K, Walz, T, Nudler, E. | | 登録日 | 2020-06-04 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (7.7 Å) | | 主引用文献 | Pre-termination Transcription Complex: Structure and Function.
Mol.Cell, 81, 2021
|
|
6XAS
 
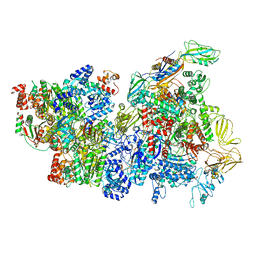 | | CryoEM Structure of E. coli Rho-dependent Transcription Pre-termination Complex | | 分子名称: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Hao, Z.T, Kim, H.K, Walz, T, Nudler, E. | | 登録日 | 2020-06-04 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Pre-termination Transcription Complex: Structure and Function.
Mol.Cell, 81, 2021
|
|
5JI7
 
 | |
5JI9
 
 | |
5JIU
 
 | |
