6K8I
 
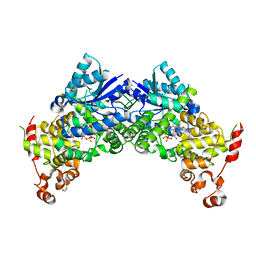 | | Crystal structure of Arabidopsis thaliana CRY2 | | 分子名称: | Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Ma, L, Wang, X, Guan, Z, Yin, P. | | 登録日 | 2019-06-12 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.697 Å) | | 主引用文献 | Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8K3K
 
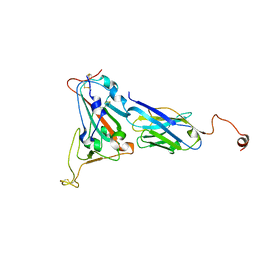 | |
8FWF
 
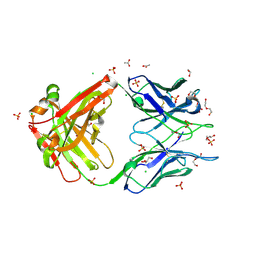 | | Crystal structure of Apo form Fab235 | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Tan, K, Kim, M, Reinherz, E.L. | | 登録日 | 2023-01-21 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
8FXJ
 
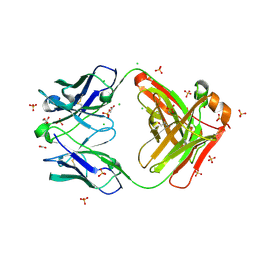 | | Crystal structure of Fab460 | | 分子名称: | ACETATE ION, CHLORIDE ION, Fab460, ... | | 著者 | Tan, K, Kim, M, Reinherz, E.L. | | 登録日 | 2023-01-24 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
8FYM
 
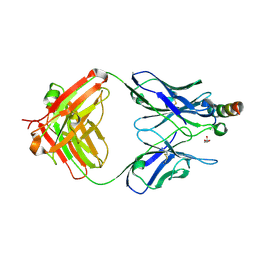 | | Crystal structure of Fab235 in complex with MPER peptide | | 分子名称: | ALA-SER-LEU-TRP-ASN-TRP-PHE-ASN-ILE-THR-ASN-TRP-LEU-TRP-TYR-ILE-LYS-LYS-LYS, CHLORIDE ION, Fab235, ... | | 著者 | Tan, K, Kim, M, Reinherz, E.L. | | 登録日 | 2023-01-26 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
8FZ2
 
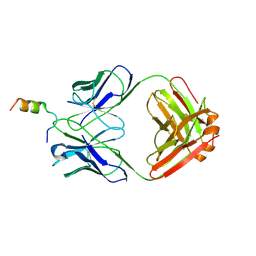 | | Crystal structure of Fab460 in complex with MPER peptide | | 分子名称: | Fab460, H chain, L chain, ... | | 著者 | Tan, K, Kim, M, Reinherz, E.L. | | 登録日 | 2023-01-27 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
6JD7
 
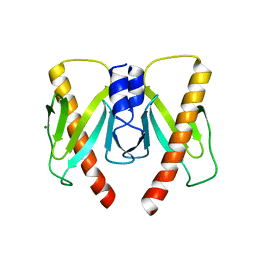 | | Crystal structure of anti-CRISPR protein AcrIIC2 dimer | | 分子名称: | ACETATE ION, AcrIIC2, MAGNESIUM ION | | 著者 | Cheng, Z, Huang, X, Sun, W, Wang, Y.L. | | 登録日 | 2019-01-31 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2.
Nat Commun, 10, 2019
|
|
8K46
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | 登録日 | 2023-07-17 | | 公開日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K45
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb4 nanobody, ... | | 著者 | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | 登録日 | 2023-07-17 | | 公開日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K47
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | 登録日 | 2023-07-17 | | 公開日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
7YFG
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine and glutamate (major class in asymmetry) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | 著者 | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | 登録日 | 2022-07-08 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFH
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine, glutamate and (R)-PYD-106 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | 著者 | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | 登録日 | 2022-07-08 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6KWY
 
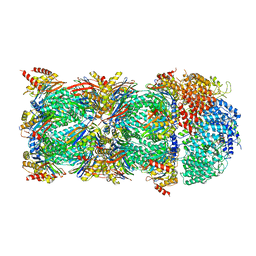 | | human PA200-20S complex | | 分子名称: | INOSITOL HEXAKISPHOSPHATE, Proteasome activator complex subunit 4, Proteasome subunit alpha type-1, ... | | 著者 | Ouyang, S, Hongxin, G. | | 登録日 | 2019-09-09 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.72 Å) | | 主引用文献 | Cryo-EM structures of the human PA200 and PA200-20S complex reveal regulation of proteasome gate opening and two PA200 apertures.
Plos Biol., 18, 2020
|
|
3JCO
 
 | | Structure of yeast 26S proteasome in M1 state derived from Titan dataset | | 分子名称: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | 著者 | Luan, B, Huang, X.L, Wu, J.P, Shi, Y.G, Wang, F. | | 登録日 | 2016-01-06 | | 公開日 | 2016-06-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Structure of an endogenous yeast 26S proteasome reveals two major conformational states.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7BWJ
 
 | | crystal structure of SARS-CoV-2 antibody with RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | 著者 | Wang, X, Ge, J. | | 登録日 | 2020-04-14 | | 公開日 | 2020-06-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Human neutralizing antibodies elicited by SARS-CoV-2 infection.
Nature, 584, 2020
|
|
6OT1
 
 | |
6OSY
 
 | |
6PM9
 
 | | Crystal structure of the core catalytic domain of human O-GlcNAcase bound to MK-8719 | | 分子名称: | (3aR,5S,6S,7R,7aR)-5-(difluoromethyl)-2-(ethylamino)-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, O-GlcNAcase TIM-barrel domain, O-GlcNAcase stalk domain | | 著者 | Klein, D.J, Selnick, H.G, Duffy, J.L, McEachern, E.J. | | 登録日 | 2019-07-01 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Discovery of MK-8719, a Potent O-GlcNAcase Inhibitor as a Potential Treatment for Tauopathies.
J.Med.Chem., 62, 2019
|
|
7BV6
 
 | |
6N05
 
 | | Structure of anti-crispr protein, AcrIIC2 | | 分子名称: | AcrIIC2 | | 著者 | Shah, M, Thavalingham, A, Maxwell, K.L, Moraes, T.F. | | 登録日 | 2018-11-06 | | 公開日 | 2019-06-05 | | 最終更新日 | 2020-01-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2.
Nat Commun, 10, 2019
|
|
7T79
 
 | |
7T78
 
 | |
7BV4
 
 | | Crystal structure of STX17 LIR region in complex with GABARAP | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein, ... | | 著者 | Li, Y, Pan, L.F. | | 登録日 | 2020-04-09 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Decoding three distinct states of the Syntaxin17 SNARE motif in mediating autophagosome-lysosome fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1CLK
 
 | | CRYSTAL STRUCTURE OF STREPTOMYCES DIASTATICUS NO.7 STRAIN M1033 XYLOSE ISOMERASE AT 1.9 A RESOLUTION WITH PSEUDO-I222 SPACE GROUP | | 分子名称: | COBALT (II) ION, MAGNESIUM ION, XYLOSE ISOMERASE | | 著者 | Niu, L, Teng, M, Zhu, X, Gong, W. | | 登録日 | 1999-04-29 | | 公開日 | 2000-05-03 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of xylose isomerase from Streptomyces diastaticus no. 7 strain M1033 at 1.85 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
4FFW
 
 | | Crystal Structure of Dipeptidyl Peptidase IV (DPP4, DPP-IV, CD26) in Complex with Fab + sitagliptin | | 分子名称: | (2R)-4-OXO-4-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-A MINE, Dipeptidyl peptidase 4, Fab heavy chain, ... | | 著者 | Wang, Z, Sudom, A, Walker, N.P, Min, X. | | 登録日 | 2012-06-01 | | 公開日 | 2012-12-12 | | 最終更新日 | 2021-05-19 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | An Inhibitory Antibody Against DPP IV Improves Glucose Tolerance in vivo - Validation of Large Molecule Approach for DPP IV Inhibition
To be published
|
|
