5XWP
 
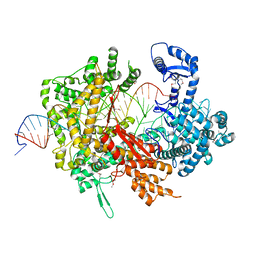 | | Crystal structure of LbuCas13a-crRNA-target RNA ternary complex | | 分子名称: | RNA (30-MER), RNA (59-MER), Uncharacterized protein | | 著者 | Liu, L, Li, X, Li, Z, Wang, Y. | | 登録日 | 2017-06-30 | | 公開日 | 2017-09-13 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.086 Å) | | 主引用文献 | The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a.
Cell, 170, 2017
|
|
3J2Z
 
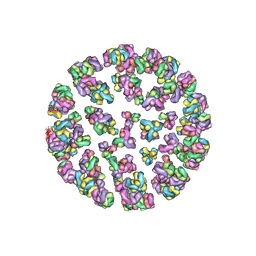 | |
3J2X
 
 | |
3J30
 
 | |
6VIK
 
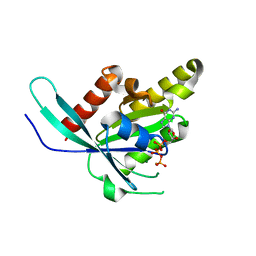 | |
8HGO
 
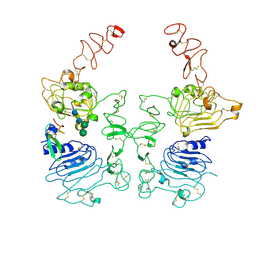 | | The EGF-bound EGFR/HER2 ectodomain complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor, ... | | 著者 | Zhang, Z, Bai, X. | | 登録日 | 2022-11-15 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-03-01 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | Structure and dynamics of the EGFR/HER2 heterodimer.
Cell Discov, 9, 2023
|
|
8HGP
 
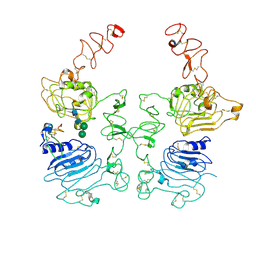 | | The EREG-bound EGFR/HER2 ectodomain complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | 著者 | Zhang, Z, Bai, X. | | 登録日 | 2022-11-15 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-03-01 | | 実験手法 | ELECTRON MICROSCOPY (4.53 Å) | | 主引用文献 | Structure and dynamics of the EGFR/HER2 heterodimer.
Cell Discov, 9, 2023
|
|
8HGS
 
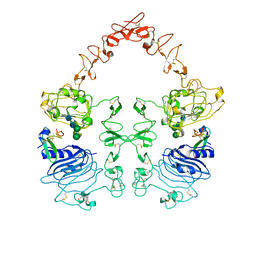 | | The EGF-bound EGFR ectodomain homodimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, Pro-epidermal growth factor, ... | | 著者 | Zhang, Z, Bai, X. | | 登録日 | 2022-11-15 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-03-01 | | 実験手法 | ELECTRON MICROSCOPY (3.81 Å) | | 主引用文献 | Structure and dynamics of the EGFR/HER2 heterodimer.
Cell Discov, 9, 2023
|
|
6VIH
 
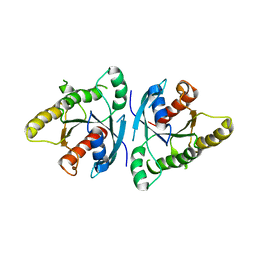 | |
4LX4
 
 | | Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase(Endo-1,4-beta-glucanase)protein | | 著者 | Dutoit, R, Delsaute, M, Berlemont, R, Van Elder, D, Galleni, M, Bauvois, C. | | 登録日 | 2013-07-29 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.556 Å) | | 主引用文献 | Crystal structure determination of Pseudomonas stutzeri A1501 endoglucanase Cel5A: the search for a molecular basis for glycosynthesis in GH5_5 enzymes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6VIJ
 
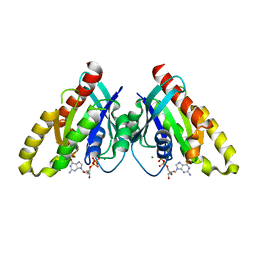 | |
6WHC
 
 | | CryoEM Structure of the glucagon receptor with a dual-agonist peptide | | 分子名称: | Dual-agonist peptide, Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Belousoff, M.J, Sexton, P, Danev, R. | | 登録日 | 2020-04-07 | | 公開日 | 2020-05-27 | | 最終更新日 | 2020-07-22 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-electron microscopy structure of the glucagon receptor with a dual-agonist peptide.
J.Biol.Chem., 295, 2020
|
|
5X4S
 
 | | Structure of the N-terminal domain (NTD)of SARS-CoV spike protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | 登録日 | 2017-02-14 | | 公開日 | 2017-05-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
6W3I
 
 | | Crystal structure of a FAM46C mutant in complex with Plk4 | | 分子名称: | Serine/threonine-protein kinase PLK4, Terminal nucleotidyltransferase 5C | | 著者 | Chen, H, Shang, G.J, Lu, D.F, Zhang, X.W. | | 登録日 | 2020-03-09 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.802 Å) | | 主引用文献 | Structural and Functional Analyses of the FAM46C/Plk4 Complex.
Structure, 28, 2020
|
|
8HF2
 
 | | Cryo-EM structure of WeiTsing | | 分子名称: | PRA1 family protein | | 著者 | Qin, L, Tang, L.H, Chen, Y.H. | | 登録日 | 2022-11-09 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (4.14 Å) | | 主引用文献 | WeiTsing, a pericycle-expressed ion channel, safeguards the stele to confer clubroot resistance.
Cell, 186, 2023
|
|
6W38
 
 | | Crystal structure of the FAM46C/Plk4 complex | | 分子名称: | Serine/threonine-protein kinase PLK4, Terminal nucleotidyltransferase 5C | | 著者 | Chen, H, Lu, D.F, Shang, G.J, Zhang, X.W. | | 登録日 | 2020-03-09 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (4.48 Å) | | 主引用文献 | Structural and Functional Analyses of the FAM46C/Plk4 Complex.
Structure, 28, 2020
|
|
220L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
225L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, PARA-XYLENE, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
228L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
226L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
223L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
8HTV
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3a | | 分子名称: | 1-(5,6-dihydrobenzo[b][1]benzazepin-11-yl)-2-sulfanyl-ethanone, 3C-like proteinase | | 著者 | Su, H.X, Nie, T.Q, Li, M.J, Xu, Y.C. | | 登録日 | 2022-12-21 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Discovery and Mechanism Study of SARS-CoV-2 3C-like Protease Inhibitors with a New Reactive Group.
J.Med.Chem., 66, 2023
|
|
222L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
229L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GUANIDINE, ... | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-26 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
227L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
