7VH5
 
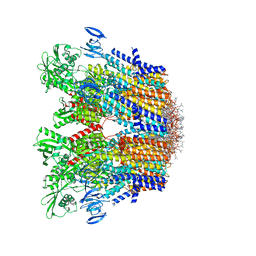 | | Cryo-EM structure of the hexameric plasma membrane H+-ATPase in the autoinhibited state (pH 7.4, C1 symmetry) | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Plasma membrane ATPase 1, SPHINGOSINE | | 著者 | Zhao, P, Zhao, C, Chen, D, Yun, C, Li, H, Bai, L. | | 登録日 | 2021-09-21 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure and activation mechanism of the hexameric plasma membrane H + -ATPase.
Nat Commun, 12, 2021
|
|
7VH6
 
 | | Cryo-EM structure of the hexameric plasma membrane H+-ATPase in the active state (pH 6.0, BeF3-, conformation 1, C1 symmetry) | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, BERYLLIUM TRIFLUORIDE ION, Plasma membrane ATPase 1 | | 著者 | Zhao, P, Zhao, C, Chen, D, Yun, C, Li, H, Bai, L. | | 登録日 | 2021-09-21 | | 公開日 | 2021-11-24 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure and activation mechanism of the hexameric plasma membrane H + -ATPase.
Nat Commun, 12, 2021
|
|
6A6B
 
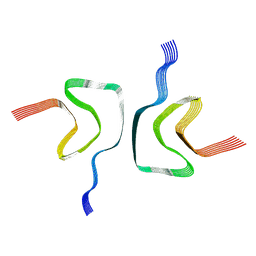 | | cryo-em structure of alpha-synuclein fiber | | 分子名称: | Alpha-synuclein | | 著者 | Li, Y.W, Zhao, C.Y, Luo, F, Liu, Z, Gui, X, Luo, Z, Zhang, X, Li, D, Liu, C, Li, X. | | 登録日 | 2018-06-27 | | 公開日 | 2018-07-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Amyloid fibril structure of alpha-synuclein determined by cryo-electron microscopy
Cell Res., 28, 2018
|
|
7PS3
 
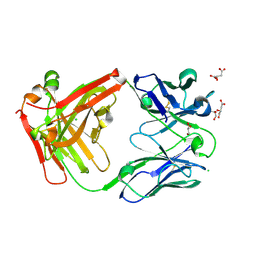 | | Crystal structure of antibody Beta-32 Fab | | 分子名称: | Beta-32 heavy chain, Beta-32 light chain, CHLORIDE ION, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-09-22 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PY2
 
 | | Structure of pathological TDP-43 filaments from ALS with FTLD | | 分子名称: | TAR DNA-binding protein 43 | | 著者 | Arseni, D, Hasegawa, H, Murzin, A.G, Kametani, F, Arai, M, Yoshida, M, Falcon, B. | | 登録日 | 2021-10-08 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.59 Å) | | 主引用文献 | Structure of pathological TDP-43 filaments from ALS with FTLD.
Nature, 601, 2022
|
|
7PS0
 
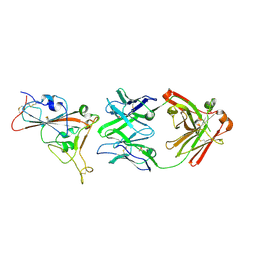 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with beta-24 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-24 heavy chain, Beta-24 light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-09-22 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PRY
 
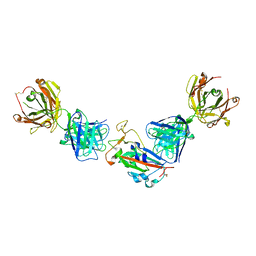 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with COVOX-45 and beta-6 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-6 Fab heavy chain, Beta-6 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-09-22 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7VOE
 
 | | Crystal structure of 5-HT2AR in complex with aripiprazole | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, 7-[4-[4-[2,3-bis(chloranyl)phenyl]piperazin-1-yl]butoxy]-3,4-dihydro-1H-quinolin-2-one, ... | | 著者 | Chen, Z, Fan, L, Wang, H, Yu, J, Lu, D, Qi, J, Nie, F, Luo, Z, Liu, Z, Cheng, J, Wang, S. | | 登録日 | 2021-10-13 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure-based design of a novel third-generation antipsychotic drug lead with potential antidepressant properties.
Nat.Neurosci., 25, 2022
|
|
7PS4
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-38 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-38 Fab heavy chain, Beta-38 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-09-22 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PS7
 
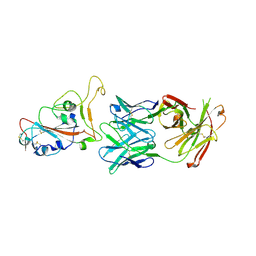 | |
7PRZ
 
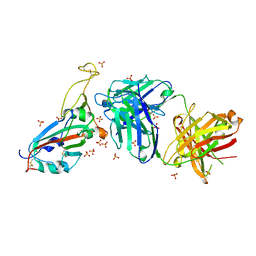 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with beta-22 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-22 Fab heavy chain, Beta-22 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-09-22 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PS2
 
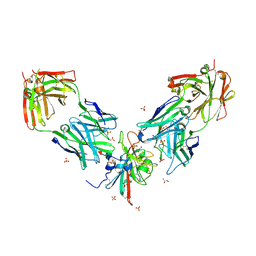 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-29 and Beta-53 Fabs | | 分子名称: | Beta-29 Fab heavy chain, Beta-29 Fab light chain, Beta-53 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-09-22 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PS5
 
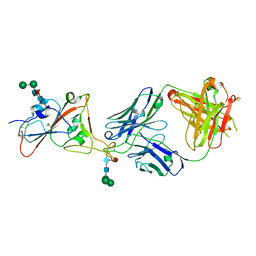 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-47 Fab | | 分子名称: | Beta-47 Fab heavy chain, Beta-47 Fab light chain, Spike protein S1, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-09-22 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.14 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7VOD
 
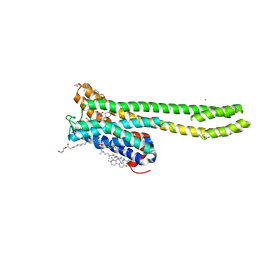 | | Crystal structure of 5-HT2AR in complex with cariprazine | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[4-[2-[4-[2,3-bis(chloranyl)phenyl]piperazin-1-yl]ethyl]cyclohexyl]-1,1-dimethyl-urea, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | 著者 | Chen, Z, Fan, L, Wang, H, Yu, J, Lu, D, Qi, J, Nie, F, Luo, Z, Liu, Z, Cheng, J, Wang, S. | | 登録日 | 2021-10-13 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structure-based design of a novel third-generation antipsychotic drug lead with potential antidepressant properties.
Nat.Neurosci., 25, 2022
|
|
7PS1
 
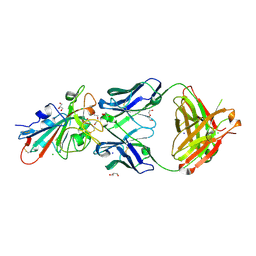 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-27 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-27 Fab heavy chain, Beta-27 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-09-22 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PS6
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-44 and Beta-54 Fabs | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, Beta-44 Fab heavy chain, Beta-44 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-09-22 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q0I
 
 | | Crystal structure of the N-terminal domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-43 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-10-14 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q0G
 
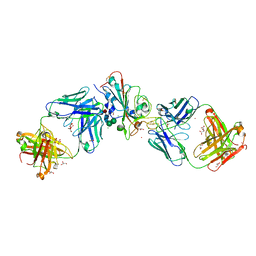 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-49 and FI-3A Fabs | | 分子名称: | Beta-49 Fab heavy chain, Beta-49 Fab light chain, CHLORIDE ION, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-10-14 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q0H
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-50 and Beta-54 | | 分子名称: | Beta-50 Fab heavy chain, Beta-50 Fab light chain, Beta-54 Fab heavy chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-10-14 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.65 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
5ZTJ
 
 | | Crystal Structure of GyraseA C-Terminal Domain from Salmonella typhi at 2.4A Resolution | | 分子名称: | DNA gyrase subunit A | | 著者 | Sachdeva, E, Gupta, D, Tiwari, P, Kaur, G, Sharma, S, Singh, T.P, Ethayathulla, A.S, Kaur, P. | | 登録日 | 2018-05-03 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The pivot point arginines identified in the beta-pinwheel structure of C-terminal domain from Salmonella Typhi DNA Gyrase A subunit.
Sci Rep, 10, 2020
|
|
5ZUG
 
 | | Structure of the bacterial acetate channel SatP | | 分子名称: | Succinate-acetate/proton symporter SatP, nonyl beta-D-glucopyranoside | | 著者 | Sun, P.C, Li, J.L, Xiao, Q.J, Guan, Z.Y, Deng, D. | | 登録日 | 2018-05-07 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.802 Å) | | 主引用文献 | Crystal structure of the bacterial acetate transporter SatP reveals that it forms a hexameric channel.
J. Biol. Chem., 293, 2018
|
|
2FTY
 
 | |
7VB6
 
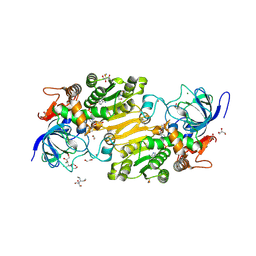 | | Crystal structure of hydroxynitrile lyase from Linum usitatissium complexed with (R)-2-hydroxy-2-methylbutanenitrile | | 分子名称: | (2R)-2-methyl-2-oxidanyl-butanenitrile, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Zheng, D, Nakabayashi, M, Asano, Y. | | 登録日 | 2021-08-30 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structural characterization of Linum usitatissimum hydroxynitrile lyase: A new cyanohydrin decomposition mechanism involving a cyano-zinc complex.
J.Biol.Chem., 298, 2022
|
|
5ZYF
 
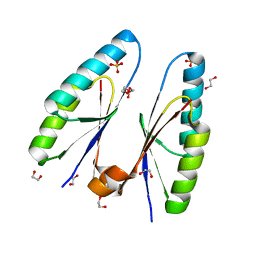 | | Crystal structure of Streptococcus pyogenes type II-A Cas2 | | 分子名称: | 1,2-ETHANEDIOL, CRISPR-associated endoribonuclease Cas2, SULFATE ION | | 著者 | Ka, D, Bae, E. | | 登録日 | 2018-05-24 | | 公開日 | 2018-08-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Molecular organization of the type II-A CRISPR adaptation module and its interaction with Cas9 via Csn2
Nucleic Acids Res., 46, 2018
|
|
7VB3
 
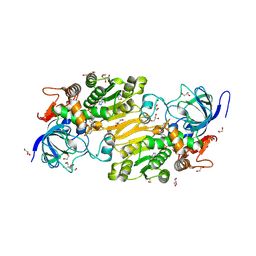 | | Crystal structure of hydroxynitrile lyase from Linum usitatissimum | | 分子名称: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aliphatic (R)-hydroxynitrile lyase, ... | | 著者 | Zheng, D, Nakabayashi, M, Asano, Y. | | 登録日 | 2021-08-30 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Structural characterization of Linum usitatissimum hydroxynitrile lyase: A new cyanohydrin decomposition mechanism involving a cyano-zinc complex.
J.Biol.Chem., 298, 2022
|
|
