6V82
 
 | | Crystal structure of tryptophan synthase from Chlamydia trachomatis D/UW-3/CX | | 分子名称: | SULFATE ION, Tryptophan synthase alpha chain, Tryptophan synthase beta chain, ... | | 著者 | Michalska, K, Maltseva, N, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-12-10 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.424 Å) | | 主引用文献 | Catalytically impaired TrpA subunit of tryptophan synthase from Chlamydia trachomatis is an allosteric regulator of TrpB.
Protein Sci., 30, 2021
|
|
6V6N
 
 | | The crystal structure of a class D beta-lactamase from Agrobacterium tumefaciens | | 分子名称: | Beta-lactamase, FORMIC ACID, GLYCEROL, ... | | 著者 | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-12-05 | | 公開日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The crystal structure of a class D beta-lactamase from Agrobacterium tumefaciens
To Be Published
|
|
5EYU
 
 | | 1.72 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-11-25 | | 公開日 | 2015-12-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | 1.72 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289
To Be Published
|
|
4X9K
 
 | | Beta-ketoacyl-acyl carrier protein synthase III-2 (FabH2)(C113A) from Vibrio cholerae | | 分子名称: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2, GLYCEROL, MALONATE ION, ... | | 著者 | Hou, J, Chruszcz, M, Zheng, H, Grabowski, M, Chordia, M.D, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-12-11 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Structural and enzymatic studies of beta-ketoacyl-(acyl carrier protein) synthase III (FabH) from Vibrio cholerae
to be published
|
|
5EWQ
 
 | | The crystal structure of an amidase family protein from Bacillus anthracis str. Ames | | 分子名称: | ACETATE ION, Amidase | | 著者 | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-11-20 | | 公開日 | 2015-12-09 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | The crystal structure of an amidase family protein from Bacillus anthracis str. Ames
To Be Published
|
|
4QGL
 
 | | Acireductone dioxygenase from Bacillus anthracis with three cadmium ions | | 分子名称: | Acireductone dioxygenase, CADMIUM ION | | 著者 | Milaczewska, A.M, Chruszcz, M, Majorek, K.A, Porebski, P.J, Borowski, T, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-05-23 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Acireductone dioxygenase from Bacillus anthracis with three cadmium ions
To be Published
|
|
6V98
 
 | | Crystal structure of Type VI secretion system effector, TseH (VCA0285) | | 分子名称: | CALCIUM ION, Cysteine hydrolase | | 著者 | Watanabe, N, Hersch, S.J, Dong, T.G, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-12-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2020-05-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Envelope stress responses defend against type six secretion system attacks independently of immunity proteins.
Nat Microbiol, 5, 2020
|
|
6VTV
 
 | | Crystal structure of PuuD gamma-glutamyl-gamma-aminobutyrate hydrolase from E. coli | | 分子名称: | Gamma-glutamyl-gamma-aminobutyrate hydrolase PuuD, MANGANESE (II) ION | | 著者 | Stogios, P.J, EVDOKIMOVA, E, DI LEO, R, SAVCHENKO, A, JOACHIMIAK, A, SATCHELL, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-02-13 | | 公開日 | 2020-02-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | PuuD gamma-glutamyl-gamma-aminobutyrate hydrolase
To Be Published
|
|
5EZ4
 
 | | 2.11 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M/Y450L double mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-11-26 | | 公開日 | 2015-12-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | 2.11 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M/Y450L double mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289
To Be Published
|
|
6W6Y
 
 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in complex with AMP | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Non-structural protein 3 | | 著者 | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-18 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.451 Å) | | 主引用文献 | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
6W75
 
 | | 1.95 Angstrom Resolution Crystal Structure of NSP10 - NSP16 Complex from SARS-CoV-2 | | 分子名称: | 2'-O-methyltransferase, FORMIC ACID, Non-structural protein 10, ... | | 著者 | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Wiersum, G, Godzik, A, Jaroszewski, L, Stogios, P.J, Skarina, T, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-18 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.951 Å) | | 主引用文献 | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
6W81
 
 | |
6WCO
 
 | |
5F7Q
 
 | |
6WHL
 
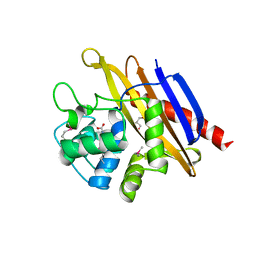 | |
6WEN
 
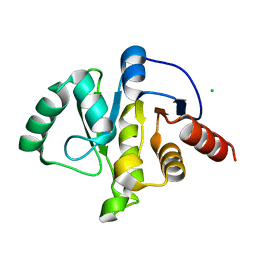 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in the apo form | | 分子名称: | CHLORIDE ION, Non-structural protein 3 | | 著者 | Michalska, K, Stols, L, Jedrzejczak, R, Endres, M, Babnigg, G, Kim, Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-02 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
6WKQ
 
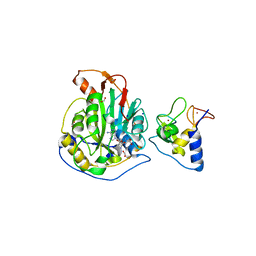 | | 1.98 Angstrom Resolution Crystal Structure of NSP16-NSP10 Heterodimer from SARS-CoV-2 in Complex with Sinefungin | | 分子名称: | 2'-O-methyltransferase, FORMIC ACID, Non-structural protein 10, ... | | 著者 | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-16 | | 公開日 | 2020-04-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
6WZU
 
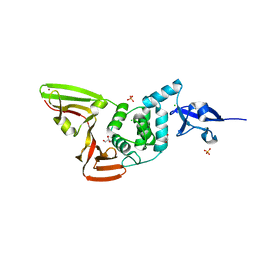 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , P3221 space group | | 分子名称: | CHLORIDE ION, GLYCEROL, Non-structural protein 3, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-05-14 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
6W79
 
 | |
5END
 
 | | Crystal structure of beta-ketoacyl-acyl carrier protein reductase (FabG)(Q152A) from Vibrio cholerae | | 分子名称: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, GLYCEROL | | 著者 | Hou, J, Cooper, D.R, Zheng, H, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-11-09 | | 公開日 | 2015-12-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
8DIL
 
 | | Crystal structure of putative nitroreductase from Salmonella enterica | | 分子名称: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Chang, C, Skarina, T, Mesa, N, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-06-29 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of putative nitroreductase from Salmonella enterica
to be published
|
|
7JH3
 
 | | Crystal structure of 4-aminobutyrate aminotransferase PuuE from Escherichia coli in complex with PLP | | 分子名称: | 4-aminobutyrate aminotransferase PuuE, DI(HYDROXYETHYL)ETHER | | 著者 | Valleau, D, Evdokimova, E, Stogios, P.J, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-20 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Crystal structure of 4-aminobutyrate aminotransferase PuuE from Escherichia coli in complex with PLP
To Be Published
|
|
7JIB
 
 | | Room Temperature Crystal Structure of Nsp10/Nsp16 from SARS-CoV-2 with Substrates and Products of 2'-O-methylation of the Cap-1 | | 分子名称: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | 著者 | Wilamowski, M, Minasov, G, Kim, Y, Sherrell, D.A, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-23 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4WH5
 
 | | Crystal structure of lincosamide antibiotic adenylyltransferase LnuA, lincomycin-bound | | 分子名称: | CHLORIDE ION, LINCOMYCIN, Lincosamide resistance protein, ... | | 著者 | Stogios, P.J, Dong, A, Minasov, G, Evdokimova, E, Egorova, O, Kudritska, M, Yim, O, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-09-20 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF LINCOSAMIDE ANTIBIOTIC ADENYLYLTRANSFERASE LNUA, LINCOMYCIN BOUND
To Be Published
|
|
6W4H
 
 | | 1.80 Angstrom Resolution Crystal Structure of NSP16 - NSP10 Complex from SARS-CoV-2 | | 分子名称: | 2'-O-methyltransferase, ACETATE ION, Non-structural protein 10, ... | | 著者 | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Wiersum, G, Godzik, A, Jaroszewski, L, Stogios, P.J, Skarina, T, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-10 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
