4JVO
 
 | | Crystal structure of the double mutant (S112A, H303A) of B.anthracis mycrocine immunity protein (MccF) with alanyl sulfamoyl adenylates | | 分子名称: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, GLYCEROL, Microcin immunity protein MccF | | 著者 | Nocek, B, Tikhonov, A, Severinov, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-03-25 | | 公開日 | 2013-06-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Crystal structure of the double mutant (S112A, H303A) of B.anthracis mycrocine immunity protein (MccF) with alanyl sulfamoyl adenylates
TO BE PUBLISHED
|
|
3PSL
 
 | | Fine-tuning the stimulation of MLL1 methyltransferase activity by a histone H3 based peptide mimetic | | 分子名称: | N-alpha acetylated form of histone H3, WD repeat-containing protein 5 | | 著者 | Avdic, V, Zhang, P, Lanouette, S, Voronova, A, Skerjanc, I, Couture, J.-F. | | 登録日 | 2010-12-01 | | 公開日 | 2010-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Fine-tuning the stimulation of MLL1 methyltransferase activity by a histone H3-based peptide mimetic.
Faseb J., 25, 2011
|
|
2VDA
 
 | | Solution structure of the SecA-signal peptide complex | | 分子名称: | MALTOPORIN, TRANSLOCASE SUBUNIT SECA | | 著者 | Gelis, I, Bonvin, A.M.J.J, Keramisanou, D, Koukaki, M, Gouridis, G, Karamanou, S, Economou, A, Kalodimos, C.G. | | 登録日 | 2007-10-01 | | 公開日 | 2007-11-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis for Signal-Sequence Recognition by the Translocase Motor Seca as Determined by NMR
Cell(Cambridge,Mass.), 131, 2007
|
|
1FL3
 
 | |
6BHP
 
 | |
7C0C
 
 | |
7C0D
 
 | |
1C4L
 
 | | SOLUTION STRUCTURE OF AN RNA DUPLEX INCLUDING A C-U BASE-PAIR | | 分子名称: | RNA (5'-R(*CP*CP*UP*GP*CP*GP*UP*CP*G)-3'), RNA (5'-R(*CP*GP*AP*CP*UP*CP*AP*GP*G)-3') | | 著者 | Tanaka, Y, Kojima, C, Yamazaki, T, Kodama, T.S, Yasuno, K, Miyashita, S, Ono, A.M, Ono, A.S, Kainosho, M, Kyogoku, Y. | | 登録日 | 1999-08-30 | | 公開日 | 2000-08-09 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of an RNA duplex including a C-U base pair.
Biochemistry, 39, 2000
|
|
6DQ4
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR GSK-J1 | | 分子名称: | 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propanoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2018-06-10 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.392 Å) | | 主引用文献 | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQ9
 
 | | Linked KDM5A JMJ Domain Bound to the Covalent Inhibitor N69 i.e. [2-((3-acrylamidophenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid] | | 分子名称: | 1,2-ETHANEDIOL, 2-{(R)-[3-(acryloylamino)phenyl][2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2018-06-10 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.748 Å) | | 主引用文献 | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQD
 
 | |
6DQ5
 
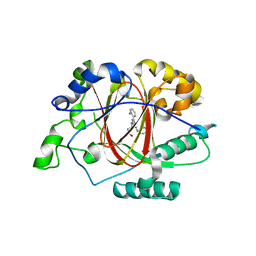 | |
6DQE
 
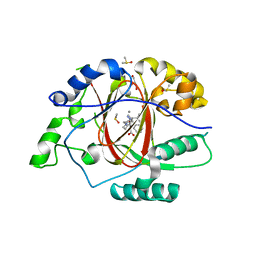 | |
1X02
 
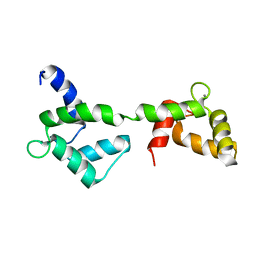 | | Solution structure of stereo array isotope labeled (SAIL) calmodulin | | 分子名称: | CALCIUM ION, calmodulin | | 著者 | Kainosho, M, Torizawa, T, Terauchi, T, Ono, A.M, Guntert, P. | | 登録日 | 2005-03-11 | | 公開日 | 2006-03-07 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Optimal isotope labelling for NMR protein structure determinations.
Nature, 440, 2006
|
|
4PE0
 
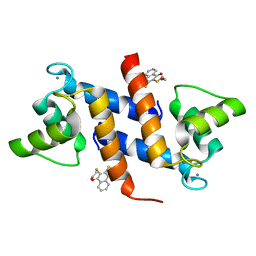 | | Crystal Structure of Calcium-loaded S100B bound to SBi4434 | | 分子名称: | 2-[(2-hydroxyethyl)sulfanyl]naphthalene-1,4-dione, CALCIUM ION, Protein S100-B | | 著者 | Cavalier, M.C, Pierce, P.D, Wilder, P.T, Neau, D, Toth, E.A, Weber, D.J. | | 登録日 | 2014-04-22 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | Covalent Small Molecule Inhibitors of Ca(2+)-Bound S100B.
Biochemistry, 53, 2014
|
|
4PDZ
 
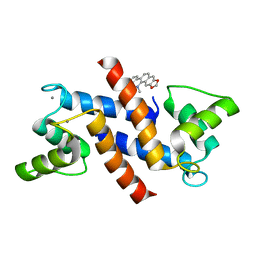 | | Crystal Structure of Calcium-loaded S100B bound to SBi4172 | | 分子名称: | 1,2-dimethoxy-12-methyl[1,3]benzodioxolo[5,6-c]phenanthridin-12-ium, CALCIUM ION, Protein S100-B | | 著者 | Cavalier, M.C, Pierce, A.D, Wilder, P.T, Neau, D, Toth, E.A, Weber, D.J. | | 登録日 | 2014-04-22 | | 公開日 | 2014-10-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Covalent Small Molecule Inhibitors of Ca(2+)-Bound S100B.
Biochemistry, 53, 2014
|
|
4PE4
 
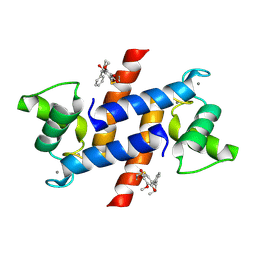 | | Crystal Structure of Calcium-loaded S100B bound to SC1475 | | 分子名称: | 2,3-dimethoxy-5-[(1S)-1-phenylpropyl]benzene-1,4-diol, CALCIUM ION, Protein S100-B | | 著者 | Cavalier, M.C, Pierce, A.D, Wilder, P.T, Neau, D, Toth, E.A, Weber, D.J. | | 登録日 | 2014-04-22 | | 公開日 | 2014-10-15 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.178 Å) | | 主引用文献 | Covalent Small Molecule Inhibitors of Ca(2+)-Bound S100B.
Biochemistry, 53, 2014
|
|
5W8J
 
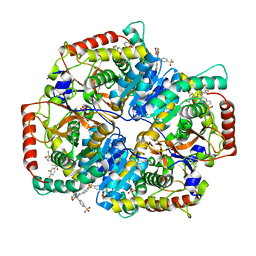 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 29 | | 分子名称: | 2-{3-(3,4-difluorophenyl)-5-hydroxy-4-[(4-sulfamoylphenyl)methyl]-1H-pyrazol-1-yl}-1,3-thiazole-4-carboxylic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Lukacs, C.M, Moulin, A. | | 登録日 | 2017-06-21 | | 公開日 | 2018-01-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
1GIP
 
 | |
5W8K
 
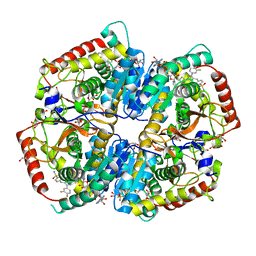 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 29 and NADH | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-{3-(3,4-difluorophenyl)-5-hydroxy-4-[(4-sulfamoylphenyl)methyl]-1H-pyrazol-1-yl}-1,3-thiazole-4-carboxylic acid, GLYCEROL, ... | | 著者 | Lukacs, C.M, Dranow, D.M. | | 登録日 | 2017-06-21 | | 公開日 | 2018-01-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
2D21
 
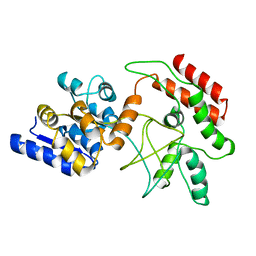 | | NMR Structure of stereo-array isotope labelled (SAIL) maltodextrin-binding protein (MBP) | | 分子名称: | Maltose-binding periplasmic protein | | 著者 | Kainosho, M, Torizawa, T, Iwashita, Y, Terauchi, T, Ono, A.M, Guntert, P. | | 登録日 | 2005-09-02 | | 公開日 | 2006-03-07 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Optimal isotope labelling for NMR protein structure determinations.
Nature, 440, 2006
|
|
2R9X
 
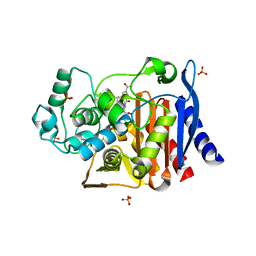 | | AmpC beta-lactamase with bound Phthalamide inhibitor | | 分子名称: | 2-[(1R)-2-carboxy-1-(naphthalen-1-ylmethyl)ethyl]-1,3-dioxo-2,3-dihydro-1H-isoindole-5-carboxylic acid, Beta-lactamase, DIMETHYL SULFOXIDE, ... | | 著者 | Babaoglu, K, Shoichet, B.K. | | 登録日 | 2007-09-13 | | 公開日 | 2008-04-15 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Comprehensive mechanistic analysis of hits from high-throughput and docking screens against beta-lactamase.
J.Med.Chem., 51, 2008
|
|
2R9W
 
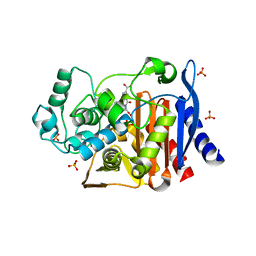 | | AmpC beta-lactamase with bound Phthalamide inhibitor | | 分子名称: | 2-[(1R)-1-carboxy-2-naphthalen-1-ylethyl]-1,3-dioxo-2,3-dihydro-1H-isoindole-5-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | 著者 | Babaoglu, K, Shoichet, B.K. | | 登録日 | 2007-09-13 | | 公開日 | 2008-04-15 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Comprehensive mechanistic analysis of hits from high-throughput and docking screens against beta-lactamase.
J.Med.Chem., 51, 2008
|
|
6RLV
 
 | | Trypanosoma brucei Seryl-tRNA Synthetase in Complex with 5'-O-(N-(L-seryl)-sulfamoyl)N3-methyluridine | | 分子名称: | 5'-O-(N-(L-seryl)-sulfamoyl)N3-methyluridine, GLYCEROL, MALONATE ION, ... | | 著者 | Pang, L, De Graef, S, Strelkov, S.V, Weeks, S.D. | | 登録日 | 2019-05-02 | | 公開日 | 2020-01-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Insights into the Binding of Natural Pyrimidine-Based Inhibitors of Class II Aminoacyl-tRNA Synthetases.
Acs Chem.Biol., 15, 2020
|
|
6S30
 
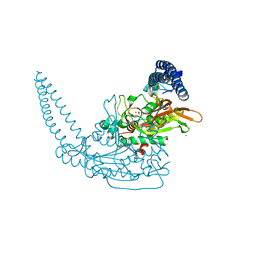 | | Crystal Structure of Seryl-tRNA Synthetase from Klebsiella pneumoniae | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Serine--tRNA ligase | | 著者 | Pang, L, De Graef, S, Weeks, S.D, Strelkov, S.V. | | 登録日 | 2019-06-23 | | 公開日 | 2020-01-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structural Insights into the Binding of Natural Pyrimidine-Based Inhibitors of Class II Aminoacyl-tRNA Synthetases.
Acs Chem.Biol., 15, 2020
|
|
