7EI8
 
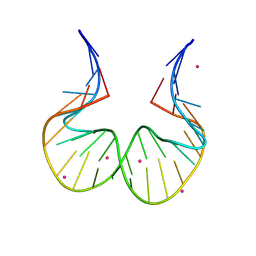 | | RNA kink-turn motif with 2-aminopurine | | 分子名称: | COBALT (II) ION, RNA (5'-R(*GP*GP*C)-D(P*(2PR)P*(2PR)P*(2PR)P*(2PR))-R(P*AP*AP*CP*CP*GP*GP*GP*GP*AP*GP*CP*C)-3') | | 著者 | Kondo, J, Saisu, S. | | 登録日 | 2021-03-30 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | RNA kink-turn motif with 2-aminopurine
To Be Published
|
|
7EIA
 
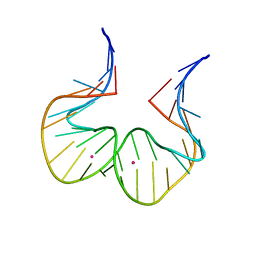 | |
7BSH
 
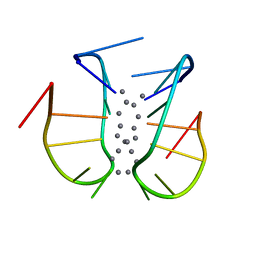 | |
6JBF
 
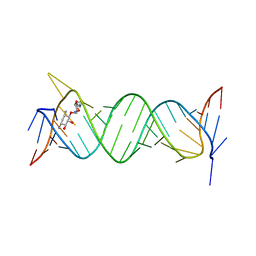 | | Crystal structure of the bacterial ribosomal decoding site in complex with 4'-deoxy-4'-fluoro neamine analog (axial 4'-F) | | 分子名称: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,4,6-trideoxy-4-fluoro-alpha-D-galactopyranoside, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | 著者 | Kanazawa, H, Auffinger, P, Hanessian, S, Kondo, J. | | 登録日 | 2019-01-25 | | 公開日 | 2020-01-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of the bacterial ribosomal decoding site in complex with 4'-deoxy-4'-fluoro neamine analog (axial 4'-F)
To Be Published
|
|
6JBG
 
 | | Crystal structure of the bacterial ribosomal decoding site in complex with 4'-deoxy-4'-fluoro neamine analog (equatorial 4'-F) | | 分子名称: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,4,6-trideoxy-4-fluoro-alpha-D-glucopyranoside, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3'), RNA (5'-R(P*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | 著者 | Kanazawa, H, Auffinger, P, Hanessian, S, Kondo, J. | | 登録日 | 2019-01-25 | | 公開日 | 2020-01-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal structure of the bacterial ribosomal decoding site in complex with 4'-deoxy-4'-fluoro neamine analog (axial 4'-F)
To Be Published
|
|
6M2P
 
 | | Crystal structure of a NIR-emitting DNA-stabilized Ag16 nanocluster (A10-deletion mutant) | | 分子名称: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*CP*AP*CP*CP*TP*AP*GP*CP*G)-3'), ... | | 著者 | Kondo, J, Cerretani, C, Vosch, T. | | 登録日 | 2020-02-28 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Removal of the A10 adenosine in a DNA-stabilized Ag16 nanocluster.
Rsc Adv, 10, 2020
|
|
6IYQ
 
 | |
6JR4
 
 | | Crystal structure of a NIR-emitting DNA-stabilized Ag16 nanocluster | | 分子名称: | CALCIUM ION, DNA (5'-D(*CP*AP*CP*CP*TP*AP*GP*CP*GP*A)-3'), SILVER ION | | 著者 | Kondo, J, Cerretani, C, Kanazawa, H, Vosch, T. | | 登録日 | 2019-04-02 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.798 Å) | | 主引用文献 | Crystal structure of a NIR-Emitting DNA-Stabilized Ag16Nanocluster.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
7XKM
 
 | |
7XLW
 
 | |
7XLV
 
 | | Crystal structure of a NIR-emitting DNA-stabilized Ag16 nanocluster (G7I mutant) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*CP*AP*CP*CP*TP*AP*IP*CP*GP*A)-3'), SILVER ION | | 著者 | Kondo, J, Cerretani, C, Vosch, T. | | 登録日 | 2022-04-23 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | The effect of inosine on the spectroscopic properties and crystal structure of a NIR-emitting DNA-stabilized silver nanocluster.
Nanoscale Adv, 4, 2022
|
|
7Y8P
 
 | | Crystal structure of 4'-selenoRNA duplex | | 分子名称: | COBALT HEXAMMINE(III), RNA (5'-R(*GP*GP*AP*(IKS)P*(ILK)P*(IKS)P*GP*AP*GP*UP*CP*C)-3') | | 著者 | Kondo, J, Minakawa, N, Ohta, M, Takahashi, H, Tarashima, N. | | 登録日 | 2022-06-24 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Synthesis and properties of fully-modified 4'-selenoRNA, an endonuclease-resistant RNA analog.
Bioorg.Med.Chem., 76, 2022
|
|
7YGP
 
 | | DNA duplex containing 5OHU-T base pairs | | 分子名称: | COBALT (II) ION, COBALT HEXAMMINE(III), DNA (5'-D(*GP*GP*AP*CP*CP*TP*(OHU)P*GP*GP*TP*CP*C)-3') | | 著者 | Kondo, J, Torigoe, H, Arakawa, F. | | 登録日 | 2022-07-12 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.997 Å) | | 主引用文献 | Specific binding of Hg 2+ to mismatched base pairs involving 5-hydroxyuracil in duplex DNA.
J.Inorg.Biochem., 241, 2023
|
|
7YGO
 
 | |
7BSG
 
 | |
7BSF
 
 | |
7BSE
 
 | |
2RT8
 
 | | Structure of metallo-dna in solution | | 分子名称: | DNA (5'-D(*CP*GP*CP*GP*TP*TP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*TP*TP*CP*GP*CP*G)-3'), MERCURY (II) ION | | 著者 | Yamaguchi, H, Sebera, J, Kondo, J, Oda, S, Komuro, T, Kawamura, T, Dairaku, T, Kondo, Y, Okamoto, I, Ono, A, Burda, J.V, Kojima, C, Sychrovsky, V, Tanaka, Y. | | 登録日 | 2013-06-18 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structure of metallo-DNA with consecutive thymine-HgII-thymine base pairs explains positive entropy for the metallo base pair formation.
Nucleic Acids Res., 42, 2014
|
|
2RVP
 
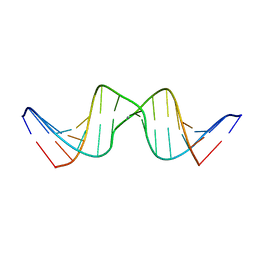 | | Solution structure of DNA Containing Metallo-Base-Pair | | 分子名称: | DNA (5'-D(*TP*AP*AP*TP*AP*TP*AP*CP*TP*TP*AP*AP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*TP*TP*AP*AP*CP*TP*AP*TP*AP*TP*TP*A)-3'), SILVER ION | | 著者 | Dairaku, T, Furuita, K, Sato, H, Sebera, J, Nakashima, K, Kondo, J, Yamanaka, D, Kondo, Y, Okamoto, I, Ono, A, Sychrovsky, V, Kojima, C, Tanaka, Y. | | 登録日 | 2016-03-22 | | 公開日 | 2016-08-31 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure Determination of an Ag(I) -Mediated Cytosine-Cytosine Base Pair within DNA Duplex in Solution with (1) H/(15) N/(109) Ag NMR Spectroscopy.
Chemistry, 22, 2016
|
|
6IUE
 
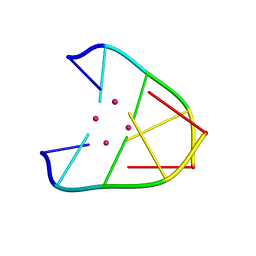 | | DNA helical wire containing Hg(II) | | 分子名称: | DNA (5'-D(*TP*TP*TP*GP*C)-3'), MERCURY (II) ION | | 著者 | Ono, A, Kanazawa, H, Ito, H, Goto, M, Nakamura, K, Saneyoshi, H, Kondo, J. | | 登録日 | 2018-11-28 | | 公開日 | 2019-10-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | A Novel DNA Helical Wire Containing HgII-Mediated T:T and T:G Pairs.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
4K31
 
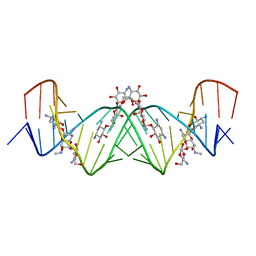 | | Crystal structure of apramycin bound to the leishmanial rRNA A-site | | 分子名称: | APRAMYCIN, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*UP*UP*CP*CP*GP*GP*AP*AP*AP*AP*GP*UP*CP*GP*C)-3') | | 著者 | Shalev, M, Kondo, J, Adir, N, Baasov, T. | | 登録日 | 2013-04-10 | | 公開日 | 2013-07-31 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.415 Å) | | 主引用文献 | Identification of the molecular attributes required for aminoglycoside activity against Leishmania.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4K32
 
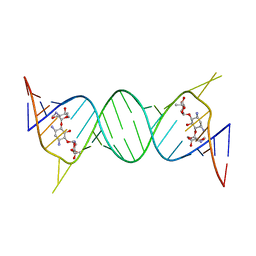 | | Crystal structure of geneticin bound to the leishmanial rRNA A-site | | 分子名称: | GENETICIN, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*UP*UP*CP*CP*GP*GP*AP*AP*AP*AP*GP*UP*CP*GP*C)-3') | | 著者 | Shalev, M, Kondo, J, Adir, N, Baasov, T. | | 登録日 | 2013-04-10 | | 公開日 | 2013-07-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Identification of the molecular attributes required for aminoglycoside activity against Leishmania.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5XWG
 
 | |
5Z1I
 
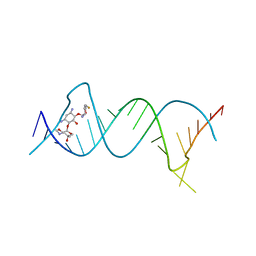 | | Crystal structure of the protozoal cytoplasmic ribosomal decoding site in complex with 6'-fluoro sisomicin | | 分子名称: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(fluoromethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, RNA (5'-R(P*GP*CP*GP*UP*CP*GP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | 著者 | Kanazawa, H, Hanessian, S, Kondo, J. | | 登録日 | 2017-12-26 | | 公開日 | 2018-05-30 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.903 Å) | | 主引用文献 | Structure-Based Design of a Eukaryote-Selective Antiprotozoal Fluorinated Aminoglycoside.
ChemMedChem, 13, 2018
|
|
5Z71
 
 | |
