7FH0
 
 | |
4EI4
 
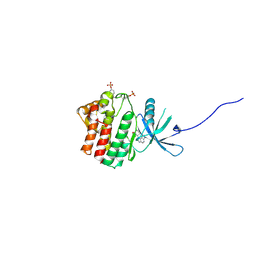 | | JAK1 kinase (JH1 domain) in complex with compound 20 | | 分子名称: | (1R,3R)-3-(2-methylimidazo[4,5-d]pyrrolo[2,3-b]pyridin-1(8H)-yl)cyclohexanol, Tyrosine-protein kinase JAK1 | | 著者 | Eigenbrot, C, Steffek, M. | | 登録日 | 2012-04-04 | | 公開日 | 2012-07-04 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Discovery and optimization of C-2 methyl imidazopyrrolopyridines as potent and orally bioavailable JAK1 inhibitors with selectivity over JAK2.
J.Med.Chem., 55, 2012
|
|
4EHZ
 
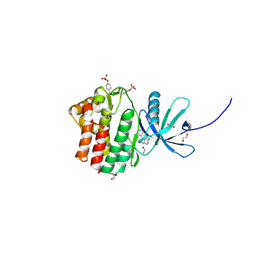 | | The Jak1 kinase domain in complex with inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2-methyl-1-(piperidin-4-yl)-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridine, Tyrosine-protein kinase JAK1 | | 著者 | Lupardus, P.J, Steffek, M. | | 登録日 | 2012-04-04 | | 公開日 | 2012-07-04 | | 最終更新日 | 2013-01-23 | | 実験手法 | X-RAY DIFFRACTION (2.174 Å) | | 主引用文献 | Discovery and optimization of C-2 methyl imidazopyrrolopyridines as potent and orally bioavailable JAK1 inhibitors with selectivity over JAK2.
J.Med.Chem., 55, 2012
|
|
4F09
 
 | |
4F08
 
 | |
5ZGM
 
 | | Crystal Structure of Parvalbumin SPVI, the Major Allergens in Mustelus griseus | | 分子名称: | CALCIUM ION, Parvalbumin SPVI, SULFATE ION | | 著者 | Yang, R.Q, Chen, Y.L, Jin, T.C, Cao, M.J. | | 登録日 | 2018-03-09 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.399 Å) | | 主引用文献 | Purification, Characterization, and Crystal Structure of Parvalbumins, the Major Allergens in Mustelus griseus.
J. Agric. Food Chem., 66, 2018
|
|
6AG0
 
 | | The X-ray Crystallographic Structure of Maltooligosaccharide-forming Amylase from Bacillus stearothermophilus STB04 | | 分子名称: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-amylase, CALCIUM ION | | 著者 | Li, Z.F, Li, Y.L, Ban, X.F, Zhang, C.Y, Jin, T.C, Xie, X.F, Gu, Z.B, Li, C.M. | | 登録日 | 2018-08-09 | | 公開日 | 2018-10-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of a maltooligosaccharide-forming amylase from Bacillus stearothermophilus STB04.
Int.J.Biol.Macromol., 138, 2019
|
|
5ZH6
 
 | | Crystal structure of Parvalbumin SPV-II of Mustelus griseus | | 分子名称: | CALCIUM ION, Parvalbumin SPV-II, SULFATE ION | | 著者 | Yang, R.Q, Chen, Y.L, Jin, T.C, Cao, M.J. | | 登録日 | 2018-03-11 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Purification, Characterization, and Crystal Structure of Parvalbumins, the Major Allergens in Mustelus griseus.
J. Agric. Food Chem., 66, 2018
|
|
5ZHQ
 
 | |
6JLC
 
 | |
6JQB
 
 | | The structure of maltooligosaccharide-forming amylase from Pseudomonas saccharophila STB07 with pseudo-maltoheptaose | | 分子名称: | 1,2-ETHANEDIOL, ACARBOSE DERIVED HEPTASACCHARIDE, CALCIUM ION, ... | | 著者 | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | 登録日 | 2019-03-30 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.101 Å) | | 主引用文献 | Structure of maltotetraose-forming amylase from Pseudomonas saccharophila STB07 provides insights into its product specificity.
Int.J.Biol.Macromol., 154, 2020
|
|
6KI0
 
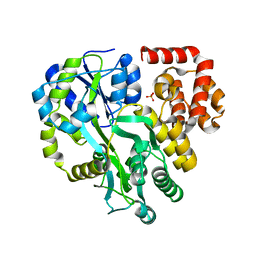 | | Crystal Structure of Human ASC-CARD | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Xu, Z.H, Jin, T.C. | | 登録日 | 2019-07-16 | | 公開日 | 2020-07-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Homotypic CARD-CARD interaction is critical for the activation of NLRP1 inflammasome.
Cell Death Dis, 12, 2021
|
|
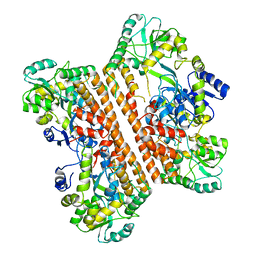
6KXG
 
 | |
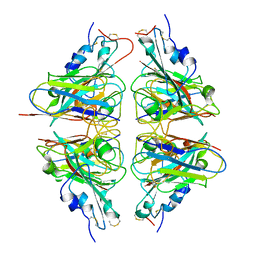
7X4I
 
 | |
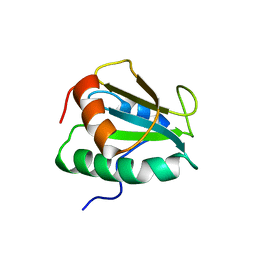
2KP1
 
 | |
2KP2
 
 | |
6X5A
 
 | | The mouse cGAS catalytic domain binding to human nucleosome that purified from HEK293T cells | | 分子名称: | Cyclic GMP-AMP synthase, DNA (natural), Histone H2A type 1, ... | | 著者 | Pengbiao, X, Pingwei, L, Baoyu, Z. | | 登録日 | 2020-05-25 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.36 Å) | | 主引用文献 | The molecular basis of tight nuclear tethering and inactivation of cGAS.
Nature, 587, 2020
|
|
6X59
 
 | | The mouse cGAS catalytic domain binding to human assembled nucleosome | | 分子名称: | Cyclic GMP-AMP synthase, DNA, Histone H2A type 1, ... | | 著者 | Pengbiao, X, Pingwei, L, Baoyu, Z. | | 登録日 | 2020-05-25 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | The molecular basis of tight nuclear tethering and inactivation of cGAS.
Nature, 587, 2020
|
|
6XJD
 
 | | Two mouse cGAS catalytic domain binding to human assembled nucleosome | | 分子名称: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1, ... | | 著者 | Xu, P, Li, P, Zhao, B. | | 登録日 | 2020-06-23 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (6.8 Å) | | 主引用文献 | The molecular basis of tight nuclear tethering and inactivation of cGAS.
Nature, 587, 2020
|
|
4NKG
 
 | | Crystal structure of SspH1 LRR domain in complex PKN1 HR1b domain | | 分子名称: | E3 ubiquitin-protein ligase sspH1, HEXANE-1,6-DIOL, Serine/threonine-protein kinase N1 | | 著者 | Keszei, A.F.A, Xiaojing, T, Mccormick, C, Zeqiraj, E, Rohde, J.R, Tyers, M, Sicheri, F. | | 登録日 | 2013-11-12 | | 公開日 | 2013-12-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure of an SspH1-PKN1 Complex Reveals the Basis for Host Substrate Recognition and Mechanism of Activation for a Bacterial E3 Ubiquitin Ligase.
Mol.Cell.Biol., 34, 2014
|
|
4NKH
 
 | | Crystal structure of SspH1 LRR domain | | 分子名称: | E3 ubiquitin-protein ligase sspH1 | | 著者 | Keszei, A.F.A, Xiaojing, T, Mccormick, C, Zeqiraj, E, Rohde, J.R, Tyers, M, Sicheri, F. | | 登録日 | 2013-11-12 | | 公開日 | 2013-12-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure of an SspH1-PKN1 Complex Reveals the Basis for Host Substrate Recognition and Mechanism of Activation for a Bacterial E3 Ubiquitin Ligase.
Mol.Cell.Biol., 34, 2014
|
|
2DJJ
 
 | |
6LOX
 
 | | Crystal Structure of human glutaminase with macrocyclic inhibitor | | 分子名称: | (E)-15,22-Dioxa-4,11-diaza-5(2,5)-thiadiazola-10(3,6)-pyridazina-1,14(1,3)-dibenzenacyclodocosaphan-18-ene-3,12-dione, Glutaminase kidney isoform, mitochondrial | | 著者 | Bian, J, Li, Z, Xu, X, Wang, J, Li, L. | | 登録日 | 2020-01-07 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure-Enabled Discovery of Novel Macrocyclic Inhibitors Targeting Glutaminase 1 Allosteric Binding Site.
J.Med.Chem., 64, 2021
|
|
2DJK
 
 | |
