8GXE
 
 | | PTPN21 FERM PTP complex | | 分子名称: | CHLORIDE ION, Tyrosine-protein phosphatase non-receptor type 21 | | 著者 | Chen, L, Zheng, Y.Y, Zhou, C. | | 登録日 | 2022-09-19 | | 公開日 | 2023-09-27 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural analysis of PTPN21 reveals a dominant-negative effect of the FERM domain on its phosphatase activity.
Sci Adv, 10, 2024
|
|
8YJC
 
 | | Structure of Vibrio vulnificus MARTX cysteine protease domain C3727A | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, INOSITOL HEXAKISPHOSPHATE, Multifunctional autoprocessing repeat-in-toxin (MARTX), ... | | 著者 | Chen, L, Khan, H, Tan, L, Li, X, Zhang, G, Im, Y.J. | | 登録日 | 2024-03-01 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-08-14 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structural basis of the activation of MARTX cysteine protease domain from Vibrio vulnificus.
Plos One, 19, 2024
|
|
8YJA
 
 | | Structure of Vibrio vulnificus MARTX cysteine protease domain lacking beta-flap | | 分子名称: | INOSITOL HEXAKISPHOSPHATE, MARTX cysteine protease domain, SODIUM ION | | 著者 | Chen, L, Khan, H, Tan, L, Li, X, Zhang, G, Im, Y.J. | | 登録日 | 2024-03-01 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-08-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis of the activation of MARTX cysteine protease domain from Vibrio vulnificus.
Plos One, 19, 2024
|
|
7WWG
 
 | | Crystal structure of Saccharomyces cerevisiae Sfh2 complexed with phosphatidylinositol in an open conformation | | 分子名称: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Phosphatidylinositol transfer protein CSR1 | | 著者 | Chen, L, Tan, L, Im, Y.J. | | 登録日 | 2022-02-12 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structural basis of ligand recognition and transport by Sfh2, a yeast phosphatidylinositol transfer protein of the Sec14 superfamily.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7WWE
 
 | |
7WWD
 
 | | Crystal structure of Saccharomyces cerevisiae Sfh2 complexed with squalene | | 分子名称: | (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene, Phosphatidylinositol transfer protein CSR1 | | 著者 | Chen, L, Tan, L, Im, Y.J. | | 登録日 | 2022-02-12 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Structural basis of ligand recognition and transport by Sfh2, a yeast phosphatidylinositol transfer protein of the Sec14 superfamily.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7WVT
 
 | | Crystal structure of Saccharomyces cerevisiae Sfh2 complexed with phosphatidylinositol | | 分子名称: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Phosphatidylinositol transfer protein CSR1 | | 著者 | Chen, L, Tan, L, Im, Y.J. | | 登録日 | 2022-02-11 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural basis of ligand recognition and transport by Sfh2, a yeast phosphatidylinositol transfer protein of the Sec14 superfamily.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7VVH
 
 | |
7VVD
 
 | |
7VUO
 
 | |
3R0M
 
 | | Crystal structure of anti-HIV llama VHH antibody A12 | | 分子名称: | Llama VHH A12, SULFATE ION | | 著者 | Chen, L, McLellan, J.S, Kwon, Y.D, Schmidt, S, Wu, X, Zhou, T, Yang, Y, Zhang, B, Forsman, A, Weiss, R.A, Verrips, T, Mascola, J, Kwong, P.D. | | 登録日 | 2011-03-08 | | 公開日 | 2012-03-14 | | 最終更新日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Single-Headed Immunoglobulins Efficiently Penetrate CD4-Binding Site and Effectively Neutralize HIV-1
To be Published
|
|
3RJQ
 
 | | Crystal structure of anti-HIV llama VHH antibody A12 in complex with C186 gp120 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C186 gp120, Llama VHH A12 | | 著者 | Chen, L, McLellan, J.S, Kwon, Y.D, Schmidt, S, Wu, X, Zhou, T, Yang, Y, Zhang, B, Forsman, A, Weiss, R.A, Verrips, T, Mascola, J, Kwong, P.D. | | 登録日 | 2011-04-15 | | 公開日 | 2012-05-02 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.601 Å) | | 主引用文献 | Crystal structure of anti-HIV A12 VHH of llama antibody in complex with C1086 gp120
To be Published
|
|
3ZIF
 
 | | Cryo-EM structures of two intermediates provide insight into adenovirus assembly and disassembly | | 分子名称: | HEXON PROTEIN, PENTON PROTEIN, PIX, ... | | 著者 | Cheng, L, Huang, X, Li, X, Xiong, W, Sun, W, Yang, C, Zhang, K, Wang, Y, Liu, H, Ji, G, Sun, F, Zheng, C, Zhu, P. | | 登録日 | 2013-01-09 | | 公開日 | 2014-01-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Cryo-Em Structures of Two Bovine Adenovirus Type 3 Intermediates
Virology, 450, 2014
|
|
5Z4M
 
 | | Structure of TailorD343A with bound UTP and Mg | | 分子名称: | MAGNESIUM ION, Terminal uridylyltransferase Tailor, URIDINE 5'-TRIPHOSPHATE | | 著者 | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | 登録日 | 2018-01-11 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
5Z4D
 
 | | Structure of Tailor in complex with AGUU RNA | | 分子名称: | RNA (5'-R(*AP*GP*UP*U)-3'), Terminal uridylyltransferase Tailor | | 著者 | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | 登録日 | 2018-01-11 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.803 Å) | | 主引用文献 | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
5Z4C
 
 | | Crystal structure of Tailor | | 分子名称: | Terminal uridylyltransferase Tailor | | 著者 | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | 登録日 | 2018-01-10 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
5Z4A
 
 | | Structure of Tailor in complex with AGU RNA | | 分子名称: | RNA (5'-R(*AP*GP*U)-3'), Terminal uridylyltransferase Tailor | | 著者 | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | 登録日 | 2018-01-10 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.637 Å) | | 主引用文献 | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
5Z4J
 
 | | Structure of Tailor in complex with U4 RNA | | 分子名称: | RNA (5'-R(*UP*UP*UP*U)-3'), Terminal uridylyltransferase Tailor | | 著者 | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | 登録日 | 2018-01-11 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
7ECA
 
 | |
3IZ3
 
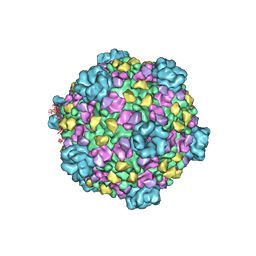 | | CryoEM structure of cytoplasmic polyhedrosis virus | | 分子名称: | Structural protein VP1, Structural protein VP3, Viral structural protein 5 | | 著者 | Cheng, L, Sun, J, Zhang, K, Mou, Z, Huang, X, Ji, G, Sun, F, Zhang, J, Zhu, P. | | 登録日 | 2010-09-14 | | 公開日 | 2011-03-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Atomic model of a cypovirus built from cryo-EM structure provides insight into the mechanism of mRNA capping.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4U1Z
 
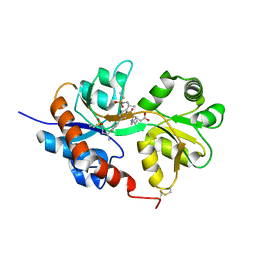 | | GluA2flip sLBD complexed with kainate and (R,R)-2b crystal form D | | 分子名称: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2,Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | 著者 | Chen, L, Gouaux, E. | | 登録日 | 2014-07-16 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.9401 Å) | | 主引用文献 | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U21
 
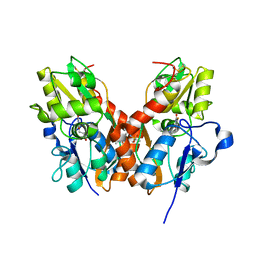 | | GluA2flip sLBD complexed with FW and (R,R)-2b crystal form E | | 分子名称: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2,Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | 著者 | Chen, L, Gouaux, E. | | 登録日 | 2014-07-16 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.3908 Å) | | 主引用文献 | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U22
 
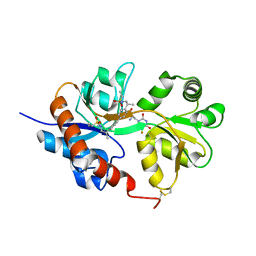 | | GluA2flip sLBD complexed with FW and (R,R)-2b crystal form D | | 分子名称: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | 著者 | Chen, L, Gouaux, E. | | 登録日 | 2014-07-16 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.4409 Å) | | 主引用文献 | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U23
 
 | | GluA2flip sLBD complexed with FW and (R,R)-2b crystal form F | | 分子名称: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2,Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | 著者 | Chen, L, Gouaux, E. | | 登録日 | 2014-07-16 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.6734 Å) | | 主引用文献 | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U1Y
 
 | | Full length GluA2-FW-(R,R)-2b complex | | 分子名称: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2, ... | | 著者 | Chen, L, Gouaux, E. | | 登録日 | 2014-07-16 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (3.8999 Å) | | 主引用文献 | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
