7CSR
 
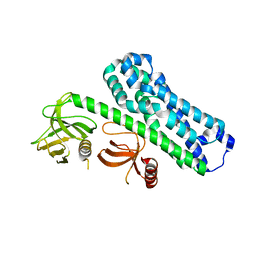 | | Structure of Ephexin4 R676L | | 分子名称: | Rho guanine nucleotide exchange factor 16 | | 著者 | Zhang, M, Lin, L, Wang, C, Zhu, J. | | 登録日 | 2020-08-17 | | 公開日 | 2021-02-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Double inhibition and activation mechanisms of Ephexin family RhoGEFs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CSO
 
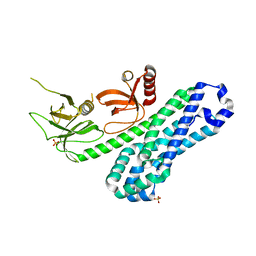 | | Structure of Ephexin4 DH-PH-SH3 | | 分子名称: | Rho guanine nucleotide exchange factor 16, SULFATE ION | | 著者 | Zhang, M, Lin, L, Wang, C, Zhu, J. | | 登録日 | 2020-08-15 | | 公開日 | 2021-02-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Double inhibition and activation mechanisms of Ephexin family RhoGEFs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1TJ2
 
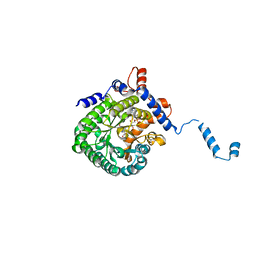 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) complexed with acetate | | 分子名称: | ACETATE ION, Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | 登録日 | 2004-06-03 | | 公開日 | 2004-10-26 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
1TJ1
 
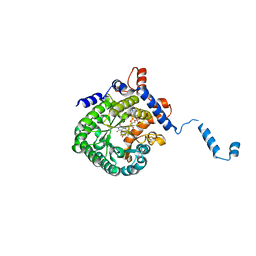 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) complexed with L-lactate | | 分子名称: | (2S)-2-HYDROXYPROPANOIC ACID, Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | 登録日 | 2004-06-03 | | 公開日 | 2004-10-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
5BT1
 
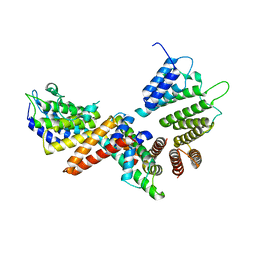 | | histone chaperone Hif1 playing with histone H2A-H2B dimer | | 分子名称: | HAT1-interacting factor 1, Histone H2A.1, Histone H2B.1 | | 著者 | Liu, H, Zhang, M, Gao, Y, Teng, M, Niu, L. | | 登録日 | 2015-06-02 | | 公開日 | 2016-10-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Structural Insights into the Association of Hif1 with Histones H2A-H2B Dimer and H3-H4 Tetramer
Structure, 24, 2016
|
|
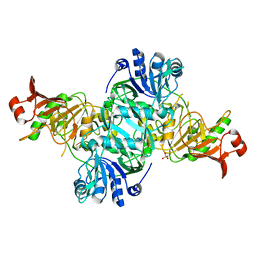
4KGB
 
 | |
3X0F
 
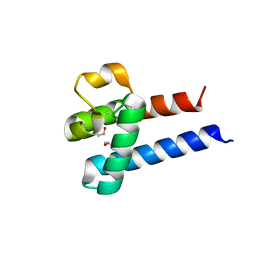 | |
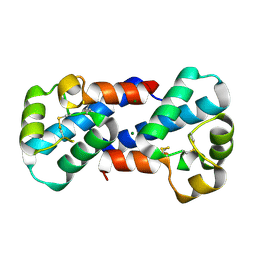
3X0E
 
 | |
3X0G
 
 | |
1TJ0
 
 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) co-crystallized with L-lactate | | 分子名称: | (2S)-2-HYDROXYPROPANOIC ACID, Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | 登録日 | 2004-06-02 | | 公開日 | 2004-10-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
1TIW
 
 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) complexed with L-Tetrahydro-2-furoic acid | | 分子名称: | Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE, TETRAHYDROFURAN-2-CARBOXYLIC ACID | | 著者 | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | 登録日 | 2004-06-02 | | 公開日 | 2004-10-26 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
7VRD
 
 | |
7V67
 
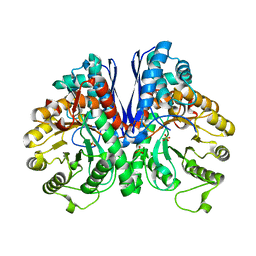 | |
5Z1F
 
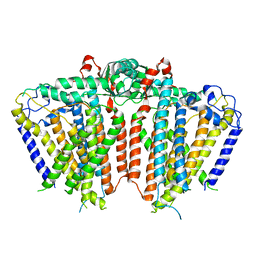 | |
5Z0Y
 
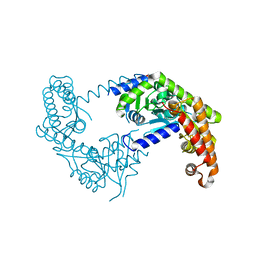 | |
6JQF
 
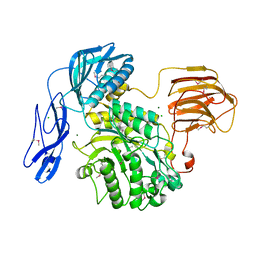 | |
8HDK
 
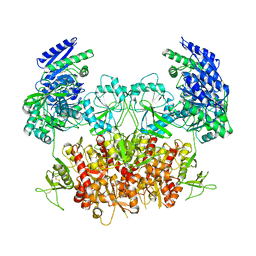 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine and glutamate (minor class in symmetry) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | 著者 | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | 登録日 | 2022-11-04 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6JPF
 
 | |
6KSF
 
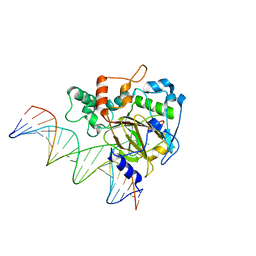 | | Crystal Structure of ALKBH1 bound to 21-mer DNA bulge | | 分子名称: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 1, CHLORIDE ION, DNA (5'-D(*DGP*DCP*DTP*DGP*DAP*DGP*DTP*DGP*DCP*DCP*DCP*DGP*DCP*DGP*DTP*DGP*DCP*DTP*DGP*DGP*DAP*DTP*DCP*DC)-3'), ... | | 著者 | Li, H, Zhang, M. | | 登録日 | 2019-08-23 | | 公開日 | 2020-04-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
7X1N
 
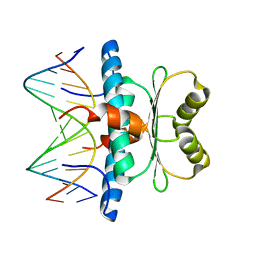 | | Crystal structure of MEF2D-MRE complex | | 分子名称: | DNA (5'-D(P*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*G)-3'), DNA (5'-D(P*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*T)-3'), Myocyte enhancer factor 2D/deleted in azoospermia associated protein 1 fusion protein | | 著者 | Zhang, H, Zhang, M, Wang, Q.Q, Chen, Z, Chen, S.J, Meng, G. | | 登録日 | 2022-02-24 | | 公開日 | 2022-05-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.315 Å) | | 主引用文献 | Functional, structural, and molecular characterizations of the leukemogenic driver MEF2D-HNRNPUL1 fusion.
Blood, 140, 2022
|
|
2LA8
 
 | |
2KBQ
 
 | |
7YFG
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine and glutamate (major class in asymmetry) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | 著者 | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | 登録日 | 2022-07-08 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFH
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine, glutamate and (R)-PYD-106 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | 著者 | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | 登録日 | 2022-07-08 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFI
 
 | | Structure of the Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA receptor in complex with glycine and glutamate | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | 著者 | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | 登録日 | 2022-07-08 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
