4J4U
 
 | | Pentamer SFTSVN | | 分子名称: | Nucleocapsid protein | | 著者 | Jiao, L, Ouyang, S, Liang, M, Niu, F, Shaw, N, Wu, W, Ding, W, Jin, C, Zhu, Y, Zhang, F, Wang, T, Li, C, Zuo, X, Luan, C.H, Li, D, Liu, Z.J. | | 登録日 | 2013-02-07 | | 公開日 | 2013-05-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.803 Å) | | 主引用文献 | Structure of severe Fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential
J.Virol., 87, 2013
|
|
4J4R
 
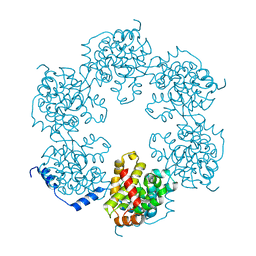 | | Hexameric SFTSVN | | 分子名称: | Nucleocapsid protein | | 著者 | Jiao, L, Ouyang, S, Liang, M, Niu, F, Shaw, N, Wu, W, Ding, W, Jin, C, Zhu, Y, Zhang, F, Wang, T, Li, C, Zuo, X, Luan, C.H, Li, D, Liu, Z.J. | | 登録日 | 2013-02-07 | | 公開日 | 2013-05-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of severe Fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential
J.Virol., 87, 2013
|
|
4J4S
 
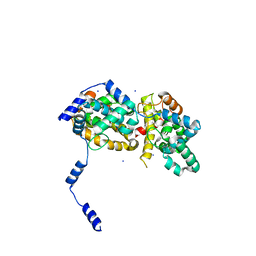 | | Triple mutant SFTAVN | | 分子名称: | Nucleocapsid protein, SODIUM ION | | 著者 | Jiao, L, Ouyang, S, Liang, M, Niu, F, Shaw, N, Wu, W, Ding, W, Jin, C, Zhu, Y, Zhang, F, Wang, T, Li, C, Zuo, X, Luan, C.H, Li, D, Liu, Z.J. | | 登録日 | 2013-02-07 | | 公開日 | 2013-05-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.442 Å) | | 主引用文献 | Structure of severe Fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential
J.Virol., 87, 2013
|
|
3ISS
 
 | | Crystal structure of enolpyruvyl-UDP-GlcNAc synthase (MurA):UDP-N-acetylmuramic acid:phosphite from Escherichia coli | | 分子名称: | PHOSPHITE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | 著者 | Jackson, S.G, Zhang, F, Chindemi, P, Junop, M.S, Berti, P.J. | | 登録日 | 2009-08-27 | | 公開日 | 2009-11-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Evidence of Kinetic Control of Ligand Binding and Staged Product Release in MurA (Enolpyruvyl UDP-GlcNAc Synthase)-Catalyzed Reactions .
Biochemistry, 48, 2009
|
|
1YKR
 
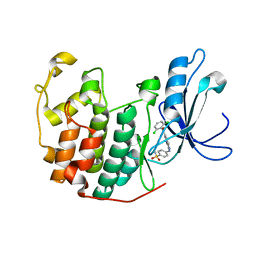 | | Crystal structure of cdk2 with an aminoimidazo pyridine inhibitor | | 分子名称: | 4-{[6-(2,6-DICHLOROBENZOYL)IMIDAZO[1,2-A]PYRIDIN-2-YL]AMINO}BENZENESULFONAMIDE, Cell division protein kinase 2 | | 著者 | Hamdouchi, C, Zhong, B, Mendoza, J, Jaramillo, C, Zhang, F, Brooks, H.B. | | 登録日 | 2005-01-18 | | 公開日 | 2006-01-24 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-based design of a new class of highly selective aminoimidazo[1,2-a]pyridine-based inhibitors of cyclin dependent kinases
Bioorg.Med.Chem.Lett., 15, 2005
|
|
9CEX
 
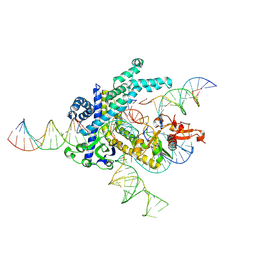 | | Spizellomyces punctatus Fanzor (SpuFz) State 4 | | 分子名称: | DNA (29-MER), DNA (5'-D(*(MG)*(MG)P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CF2
 
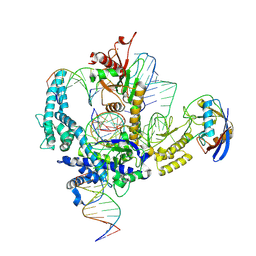 | | Parasitella parasitica Fanzor (PpFz) State 3 | | 分子名称: | DNA non-target strand, DNA substrate model, DNA target strand, ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9C0I
 
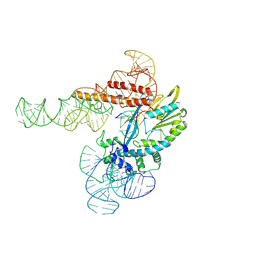 | |
8W1P
 
 | |
9CEU
 
 | | Spizellomyces punctatus Fanzor (SpuFz) State 1 | | 分子名称: | DNA (5'-D(P*CP*CP*TP*AP*TP*AP*GP*AP*TP*AP*TP*GP*CP*CP*CP*GP*GP*GP*TP*AP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), Maltose/maltodextrin-binding periplasmic protein,Spizellomyces punctatus Fanzor 1, ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CF1
 
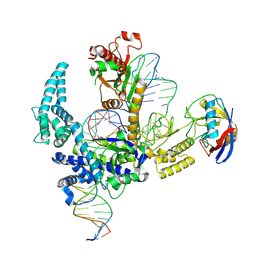 | | Parasitella parasitica Fanzor (PpFz) State 2 | | 分子名称: | DNA non-target strand, DNA target strand, Maltose/maltodextrin-binding periplasmic protein,Parasitella parasitica Fanzor 1, ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CES
 
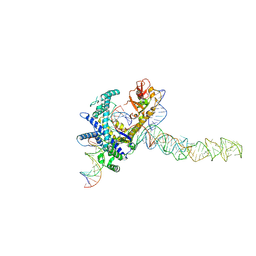 | | Guillardia theta Fanzor (GtFz) State 2 | | 分子名称: | DNA (5'-D(P*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*TP*AP*AP*AP*GP*GP*CP*CP*CP*CP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*GP*GP*GP*GP*CP*CP*TP*TP*TP*AP*AP*G)-3'), Maltose/maltodextrin-binding periplasmic protein,Guillardia theta Fanzor1, ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CEW
 
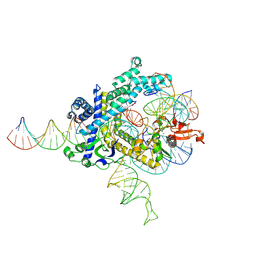 | | Spizellomyces punctatus Fanzor (SpuFz) State 3 | | 分子名称: | DNA (29-MER), DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (2.88 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CF3
 
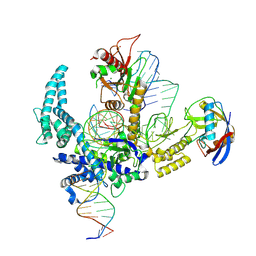 | | Parasitella parasitica Fanzor (PpFz) State 4 | | 分子名称: | DNA non-target strand, DNA target strand, MAGNESIUM ION, ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CF0
 
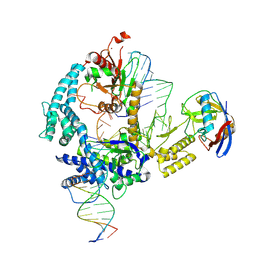 | | Parasitella parasitica Fanzor (PpFz) State 1 | | 分子名称: | DNA non-target strand, DNA target strand, Maltose/maltodextrin-binding periplasmic protein,Parasitella parasitica Fanzor 1, ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CER
 
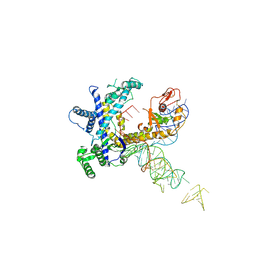 | | Guillardia theta Fanzor (GtFz) State 1 | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein,Guillardia theta Fanzor1, RNA (142-MER), ZINC ION | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CET
 
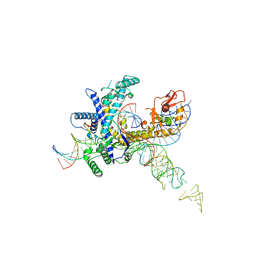 | | Guillardia theta Fanzor (GtFz) State 3 | | 分子名称: | DNA (28-MER), DNA (5'-D(P*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*TP*AP*AP*AP*GP*GP*CP*CP*CP*CP*GP*GP*G)-3'), Maltose/maltodextrin-binding periplasmic protein,Guillardia theta Fanzor1, ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CEV
 
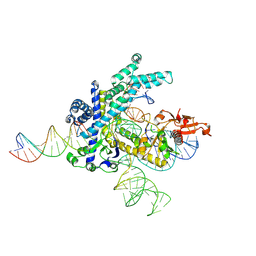 | | Spizellomyces punctatus Fanzor (SpuFz) State 2 | | 分子名称: | DNA (35-MER), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), MAGNESIUM ION, ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CEY
 
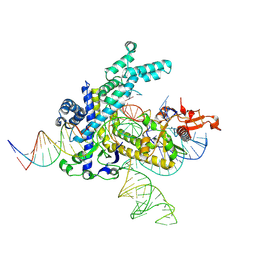 | | Spizellomyces punctatus Fanzor (SpuFz) State 5 | | 分子名称: | DNA (26-MER), DNA (36-MER), MAGNESIUM ION, ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CEZ
 
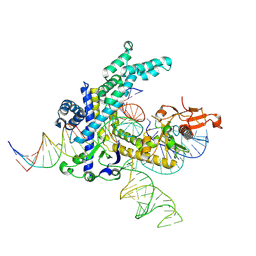 | | Spizellomyces punctatus Fanzor (SpuFz) State 6 | | 分子名称: | DNA (27-MER), DNA (5'-D(P*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), MAGNESIUM ION, ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (3.41 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
6LQX
 
 | | Crystal structure of the CBP bromodomain in complex with small molecule LC-CPin7 | | 分子名称: | (1~{S},6~{R})-6-[(1-methoxycarbonyl-3,4-dihydro-2~{H}-quinolin-6-yl)carbamoyl]cyclohex-3-ene-1-carboxylic acid, CREB-binding protein, SODIUM ION | | 著者 | Chen, Y, Zhang, F, Sun, Z, Bi, X, Luo, C. | | 登録日 | 2020-01-14 | | 公開日 | 2021-01-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Design, synthesis and biological evaluation of novel small molecule inhibitor of the CBP bromodomain with possible anti-leukemia effects
To Be Published
|
|
9J1R
 
 | | Structure of a triple-helix region of human Collagen type II from Trautec | | 分子名称: | SULFATE ION, Triple-helix region of human collagen type II | | 著者 | Fan, X, Chu, Y, Zhai, Y, Fu, S, Li, D, Cao, K, Feng, P, Wang, X, Le, H, Tang, D, Zhang, F, Qian, S. | | 登録日 | 2024-08-05 | | 公開日 | 2024-08-21 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structure of a triple-helix region of human Collagen type II from Trautec
To Be Published
|
|
8FNI
 
 | | Cryo-EM structure of RNase-treated RESC-B in trypanosomal RNA editing | | 分子名称: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | 著者 | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | 登録日 | 2022-12-27 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNK
 
 | | Cryo-EM structure of RNase-untreated RESC-B in trypanosomal RNA editing | | 分子名称: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | 著者 | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | 登録日 | 2022-12-27 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNC
 
 | | Cryo-EM structure of RNase-treated RESC-C in trypanosomal RNA editing | | 分子名称: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | 著者 | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | 登録日 | 2022-12-27 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
