8RF6
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 11A7_AL5 refined against the anomalous diffraction data | | 分子名称: | 6-iodanyl-2,3-dihydro-1,3-benzothiazol-2-amine, Host translation inhibitor nsp1 | | 著者 | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | 登録日 | 2023-12-12 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RFD
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7H2_AL2 refined against the anomalous diffraction data | | 分子名称: | (1~{R})-1-(4-iodophenyl)ethanamine, Host translation inhibitor nsp1 | | 著者 | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | 登録日 | 2023-12-12 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RF3
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7G3 refined against the anomalous diffraction data | | 分子名称: | 2-(1-benzothiophen-3-yl)ethanoic acid, Host translation inhibitor nsp1 | | 著者 | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | 登録日 | 2023-12-12 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.23 Å) | | 主引用文献 | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
5ZMC
 
 | | Structural Basis for Reactivation of -146C>T Mutant TERT Promoter by cooperative binding of p52 and ETS1/2 | | 分子名称: | DNA (5'-D(P*CP*GP*GP*GP*GP*AP*CP*CP*CP*GP*GP*AP*AP*GP*GP*G)-3'), DNA (5'-D(P*GP*CP*CP*CP*TP*TP*CP*CP*GP*GP*GP*TP*CP*CP*CP*C)-3'), Nuclear factor NF-kappa-B p100 subunit, ... | | 著者 | Xu, X, Bharath, S.R, Song, H. | | 登録日 | 2018-04-02 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Structural basis for reactivating the mutant TERT promoter by cooperative binding of p52 and ETS1.
Nat Commun, 9, 2018
|
|
7BNV
 
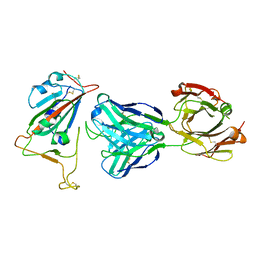 | | Crystal Structure of the SARS-CoV-2 Receptor Binding Domain in Complex with Antibody ION-300 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain, Light Chain, ... | | 著者 | Hall, G, Cowan, R, Carr, M. | | 登録日 | 2021-01-22 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Cross-Reactive SARS-CoV-2 Neutralizing Antibodies From Deep Mining of Early Patient Responses.
Front Immunol, 12, 2021
|
|
7X0F
 
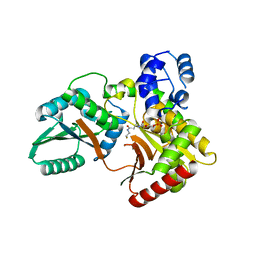 | |
7X0E
 
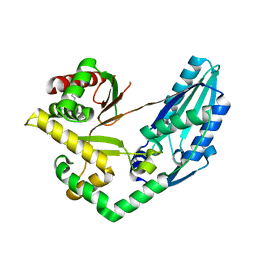 | | Structure of Pseudomonas NRPS protein, AmbB-TC in apo form | | 分子名称: | AMB antimetabolite synthase AmbB, N-methyl-N-[(2S,3R,4R,5R)-2,3,4,5,6-pentakis(oxidanyl)hexyl]nonanamide | | 著者 | ChuYuanKee, M, Bharath, S.R, Song, H. | | 登録日 | 2022-02-22 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural insights into the substrate-bound condensation domains of non-ribosomal peptide synthetase AmbB.
Sci Rep, 12, 2022
|
|
7X17
 
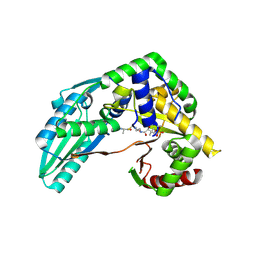 | | Structure of Pseudomonas NRPS protein, AmbB-TC bound to Ppant-L-Ala | | 分子名称: | AMB antimetabolite synthase AmbB, S-[2-[3-[[(2S)-3,3-dimethyl-2-oxidanyl-4-phosphonooxy-butanoyl]amino]propanoylamino]ethyl] (2R)-2-azanylpropanethioate | | 著者 | ChuYuanKee, M, Bharath, S.R, Song, H. | | 登録日 | 2022-02-23 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural insights into the substrate-bound condensation domains of non-ribosomal peptide synthetase AmbB.
Sci Rep, 12, 2022
|
|
5JXE
 
 | |
6SBI
 
 | | X-ray structure of murine Fumarylacetoacetate hydrolase domain containing protein 1 (FAHD1) in complex with inhibitor oxalate | | 分子名称: | Acylpyruvase FAHD1, mitochondrial, CHLORIDE ION, ... | | 著者 | Rupp, B, Naschberger, A, Weiss, A.K.H. | | 登録日 | 2019-07-21 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural and functional comparison of fumarylacetoacetate domain containing protein 1 in human and mouse.
Biosci.Rep., 40, 2020
|
|
6SBJ
 
 | | X-ray structure of mus musculus Fumarylacetoacetate hydrolase domain containing protein 1 (FAHD1) apo-form uuncomplexed | | 分子名称: | Acylpyruvase FAHD1, mitochondrial, CHLORIDE ION, ... | | 著者 | Rupp, B, Naschberger, A, Weiss, A.K.H. | | 登録日 | 2019-07-21 | | 公開日 | 2020-02-26 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Structural and functional comparison of fumarylacetoacetate domain containing protein 1 in human and mouse.
Biosci.Rep., 40, 2020
|
|
5H3U
 
 | | Sm RNA bound to GEMIN5-WD | | 分子名称: | GLYCEROL, Gem-associated protein 5, RNA (5'-R(*AP*AP*UP*UP*UP*UP*UP*GP*AP*C)-3') | | 著者 | Bharath, S.R, Tang, X, Song, H. | | 登録日 | 2016-10-27 | | 公開日 | 2016-12-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.499 Å) | | 主引用文献 | Structural basis for specific recognition of pre-snRNA by Gemin5
Cell Res., 26, 2016
|
|
5H3T
 
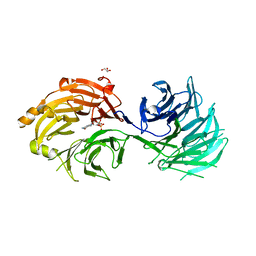 | | m7G cap bound to GEMIN5-WD | | 分子名称: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, GLYCEROL, Gem-associated protein 5 | | 著者 | Bharath, S.R, Tang, X, Song, H. | | 登録日 | 2016-10-27 | | 公開日 | 2016-12-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.571 Å) | | 主引用文献 | Structural basis for specific recognition of pre-snRNA by Gemin5
Cell Res., 26, 2016
|
|
5H3S
 
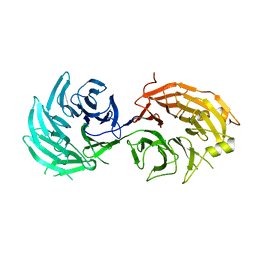 | | apo form of GEMIN5-WD | | 分子名称: | GLYCEROL, Gem-associated protein 5 | | 著者 | Bharath, S.R, Tang, X, Song, H. | | 登録日 | 2016-10-27 | | 公開日 | 2016-12-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis for specific recognition of pre-snRNA by Gemin5
Cell Res., 26, 2016
|
|
6FFT
 
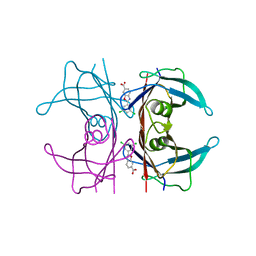 | | Neutron structure of human transthyretin (TTR) S52P mutant in complex with tafamidis at room temperature to 2A resolution (quasi-Laue) | | 分子名称: | 2-(3,5-dichlorophenyl)-1,3-benzoxazole-6-carboxylic acid, Transthyretin | | 著者 | Yee, A.W, Moulin, M, Blakeley, M.P, Haertlein, M, Mitchell, E.P, Forsyth, V.T. | | 登録日 | 2018-01-09 | | 公開日 | 2019-01-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | 主引用文献 | A molecular mechanism for transthyretin amyloidogenesis.
Nat Commun, 10, 2019
|
|
6FOG
 
 | |
6FOH
 
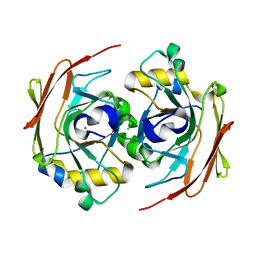 | |
5DMQ
 
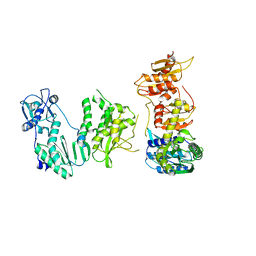 | |
5DMR
 
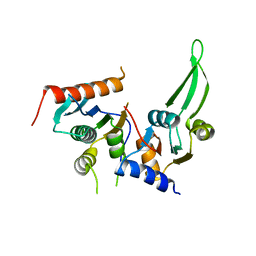 | |
4B3F
 
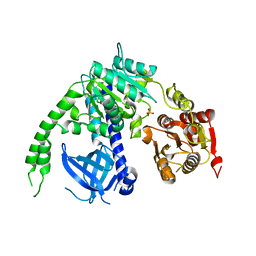 | | crystal structure of Ighmbp2 helicase | | 分子名称: | DNA-BINDING PROTEIN SMUBP-2, PHOSPHATE ION | | 著者 | Lim, S.C, Song, H. | | 登録日 | 2012-07-24 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Ighmbp2 Helicase Structure Reveals the Molecular Basis for Disease-Causing Mutations in Dmsa1.
Nucleic Acids Res., 40, 2012
|
|
4Z4R
 
 | | Crystal structure of GII.10 P domain in complex with 300mM fucose | | 分子名称: | 1,2-ETHANEDIOL, Capsid protein, beta-L-fucopyranose | | 著者 | Koromyslova, A.D, Leuthold, M.M, Hansman, G.S. | | 登録日 | 2015-04-02 | | 公開日 | 2015-05-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
4Z4T
 
 | | Crystal structure of GII.10 P domain in complex with 75mM fucose | | 分子名称: | 1,2-ETHANEDIOL, Capsid protein, NITRATE ION, ... | | 著者 | Koromyslova, A.D, Leuthold, M.M, Hansman, G.S. | | 登録日 | 2015-04-02 | | 公開日 | 2015-05-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
4Z4W
 
 | | Crystal structure of GII.10 P domain in complex with 4.7mM fucose | | 分子名称: | 1,2-ETHANEDIOL, Capsid protein, beta-L-fucopyranose | | 著者 | Koromyslova, A.D, Leuthold, M.M, Hansman, G.S. | | 登録日 | 2015-04-02 | | 公開日 | 2015-05-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
4Z4Y
 
 | | Crystal structure of GII.10 P domain in complex with 7.5mM B antigen (trisaccharide) | | 分子名称: | 1,2-ETHANEDIOL, Capsid protein, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | 著者 | Leuthold, M.M, Koromyslova, A.D, Hansman, G.S. | | 登録日 | 2015-04-02 | | 公開日 | 2015-05-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.797 Å) | | 主引用文献 | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
4Z4Z
 
 | | Crystal structure of GII.10 P domain in complex with 30mM B antigen (trisaccharide) | | 分子名称: | 1,2-ETHANEDIOL, Capsid protein, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | 著者 | Leuthold, M.M, Koromyslova, A.D, Hansman, G.S. | | 登録日 | 2015-04-02 | | 公開日 | 2015-05-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
