6IFK
 
 | | Cryo-EM structure of type III-A Csm-CTR1 complex, AMPPNP bound | | 分子名称: | CTR1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-20 | | 公開日 | 2018-12-12 | | 最終更新日 | 2019-01-23 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IFL
 
 | | Cryo-EM structure of type III-A Csm-NTR complex | | 分子名称: | NTR, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-20 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
4LDT
 
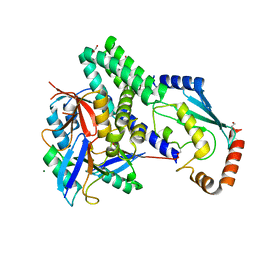 | | The structure of h/ceOTUB1-ubiquitin aldehyde-UBCH5B~Ub | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, Ubiquitin, ... | | 著者 | Wiener, R, DiBello, A.T, Lombardi, P.M, Guzzo, C.M, Zhang, X, Matunis, M.J, Wolberger, C. | | 登録日 | 2013-06-25 | | 公開日 | 2013-08-14 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6HT4
 
 | |
6IFY
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-CTR1 | | 分子名称: | CTR1, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-21 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
4MGG
 
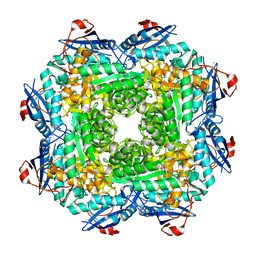 | | Crystal structure of an enolase (mandelate racemase subgroup) from labrenzia aggregata iam 12614 (target nysgrc-012903) with bound mg, space group p212121 | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, Muconate lactonizing enzyme, ... | | 著者 | Vetting, M.W, Zhang, X, Wasserman, S.R, Morisco, L.L, Sojitra, S, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-08-28 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of an enolase (mandelate racemase subgroup) from labrenzia aggregata iam 12614 (target nysgrc-012903) with bound mg, space group p212121
To be Published
|
|
6IFZ
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-CTR2-ssDNA complex | | 分子名称: | CTR2, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-21 | | 公開日 | 2018-12-12 | | 最終更新日 | 2019-01-23 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6HTS
 
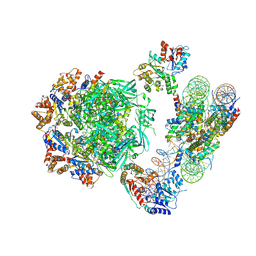 | | Cryo-EM structure of the human INO80 complex bound to nucleosome | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | 著者 | Ayala, R, Willhoft, O, Aramayo, R.J, Wilkinson, M, McCormack, E.A, Ocloo, L, Wigley, D.B, Zhang, X. | | 登録日 | 2018-10-04 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Structure and regulation of the human INO80-nucleosome complex.
Nature, 556, 2018
|
|
7WQV
 
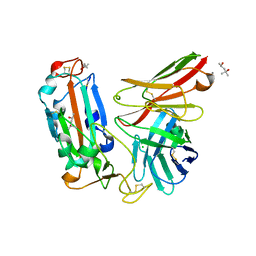 | | Crystal structure of a neutralizing monoclonal antibody (Ab08) in complex with SARS-CoV-2 receptor-binding domain (RBD) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ab08, ... | | 著者 | Zha, J, Meng, L, Zhang, X, Li, D. | | 登録日 | 2022-01-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A Spike-destructing human antibody effectively neutralizes Omicron-included SARS-CoV-2 variants with therapeutic efficacy.
Plos Pathog., 19, 2023
|
|
7XSN
 
 | | Native Tetrahymena ribozyme conformation | | 分子名称: | RNA (387-MER) | | 著者 | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | 登録日 | 2022-05-14 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSL
 
 | | Misfolded Tetrahymena ribozyme conformation 2 | | 分子名称: | RNA (388-MER) | | 著者 | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | 登録日 | 2022-05-14 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.84 Å) | | 主引用文献 | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSM
 
 | | Misfolded Tetrahymena ribozyme conformation 3 | | 分子名称: | RNA (388-MER) | | 著者 | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | 登録日 | 2022-05-14 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (4.01 Å) | | 主引用文献 | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSK
 
 | | Misfolded Tetrahymena ribozyme conformation 1 | | 分子名称: | RNA (388-MER) | | 著者 | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | 登録日 | 2022-05-14 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.53 Å) | | 主引用文献 | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6IG0
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-CTR1, ATP bound | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CTR1, MAGNESIUM ION, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-21 | | 公開日 | 2018-12-12 | | 最終更新日 | 2019-01-23 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
7W5S
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE (II) ION, ... | | 著者 | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | 登録日 | 2021-11-30 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
7W5T
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | 分子名称: | 2-OXOGLUTARIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | 登録日 | 2021-11-30 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
7W5V
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | 著者 | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | 登録日 | 2021-11-30 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
7YG8
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 5 undergoing the second-step self-splicing | | 分子名称: | MAGNESIUM ION, RNA (387-MER), RNA (5'-R(*CP*CP*CP*UP*C)-3'), ... | | 著者 | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | 登録日 | 2022-07-11 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YG9
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 1 undergoing the second-step self-splicing | | 分子名称: | MAGNESIUM ION, RNA (391-MER), RNA (5'-R(*CP*CP*CP*UP*CP*U)-3'), ... | | 著者 | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | 登録日 | 2022-07-11 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YGC
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 4 undergoing the second-step self-splicing | | 分子名称: | MAGNESIUM ION, RNA (393-MER), RNA (5'-R(*CP*CP*CP*UP*CP*UP*UP*AP*AP*CP*C)-3'), ... | | 著者 | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | 登録日 | 2022-07-11 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.65 Å) | | 主引用文献 | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YGA
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 2 undergoing the second-step self-splicing | | 分子名称: | MAGNESIUM ION, RNA (393-MER), RNA (5'-R(*CP*CP*CP*UP*CP*U)-3'), ... | | 著者 | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | 登録日 | 2022-07-11 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.35 Å) | | 主引用文献 | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YGB
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 3 undergoing the second-step self-splicing | | 分子名称: | MAGNESIUM ION, RNA (393-MER), RNA (5'-R(*CP*CP*CP*UP*CP*UP*UP*AP*AP*CP*C)-3'), ... | | 著者 | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | 登録日 | 2022-07-11 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.62 Å) | | 主引用文献 | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YGD
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 6 undergoing the second-step self-splicing | | 分子名称: | MAGNESIUM ION, RNA (384-MER), RNA (5'-R(*CP*C)-3'), ... | | 著者 | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | 登録日 | 2022-07-11 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.41 Å) | | 主引用文献 | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
6KA4
 
 | | Cryo-EM structure of the AtMLKL3 tetramer | | 分子名称: | F22L4.1 protein | | 著者 | Lisa, M, Huang, M, Zhang, X, Ryohei, T.N, Leila, B.K, Isabel, M.L.S, Florence, J, Viera, K, Dmitry, L, Jane, E.P, James, M.M, Kay, H, Paul, S.L, Chai, J, Takaki, M. | | 登録日 | 2019-06-20 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structure of the AtMLKL3 tetramer
To Be Published
|
|
4OU1
 
 | | Crystal structure of a computationally designed retro-aldolase covalently bound to folding probe 1 [(6-methoxynaphthalen-2-yl)(oxiran-2-yl)methanol] | | 分子名称: | (1S,2S)-1-(6-methoxynaphthalen-2-yl)propane-1,2-diol, BENZOIC ACID, PHOSPHATE ION, ... | | 著者 | Bhabha, G, Zhang, X, Ekiert, D.C. | | 登録日 | 2014-02-14 | | 公開日 | 2014-03-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Small molecule probes to quantify the functional fraction of a specific protein in a cell with minimal folding equilibrium shifts.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
