8HC0
 
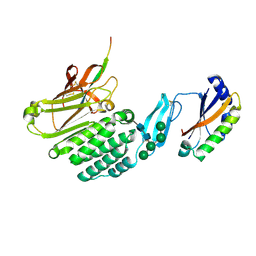 | | Crystal structure of the extracellular domains of GPR110 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G-protein coupled receptor F1, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Wang, F.F, Song, G.J. | | 登録日 | 2022-11-01 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal Structure of the Extracellular Domains of GPR110.
J.Mol.Biol., 435, 2023
|
|
7BVF
 
 | | Cryo-EM structure of Mycobacterium tuberculosis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | 分子名称: | CALCIUM ION, CARDIOLIPIN, Ethambutol, ... | | 著者 | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | 登録日 | 2020-04-10 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
5QC4
 
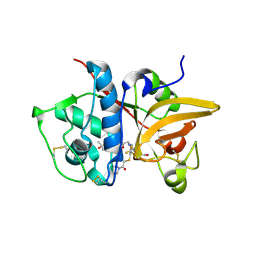 | | Crystal structure of human Cathepsin-S with bound ligand | | 分子名称: | 2-[5-[5-ethanoyl-1-[(2~{R})-2-oxidanyl-3-[4-(2-oxidanylpropan-2-yl)piperidin-1-yl]propyl]-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-3-yl]-2-(trifluoromethyl)phenyl]sulfanyl-1-pyrrolidin-1-yl-ethanone, Cathepsin S | | 著者 | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | 登録日 | 2017-08-04 | | 公開日 | 2017-12-20 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Thioether acetamides as P3 binding elements for tetrahydropyrido-pyrazole cathepsin S inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7XML
 
 | |
7BYO
 
 | |
7BYP
 
 | |
7D04
 
 | |
7D05
 
 | |
7D02
 
 | |
7D01
 
 | |
7VPR
 
 | |
7VPU
 
 | |
7VPS
 
 | |
7VPT
 
 | |
7VLN
 
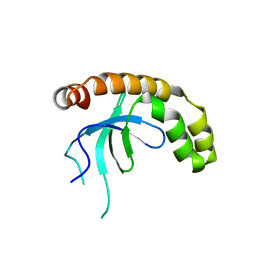 | | NSD2-PWWP1 domain bound with an imidazol-5-yl benzonitrile compound | | 分子名称: | 4-[5-[4-(aminomethyl)-2,6-dimethoxy-phenyl]-3-methyl-imidazol-4-yl]benzenecarbonitrile, Histone-lysine N-methyltransferase NSD2 | | 著者 | Cao, D.Y, Li, Y.L, Li, J, Xiong, B. | | 登録日 | 2021-10-05 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Structure-Based Discovery of a Series of NSD2-PWWP1 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7CKF
 
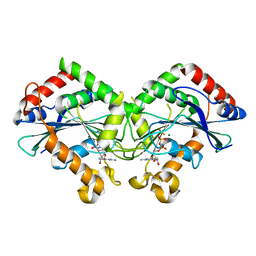 | |
7CUT
 
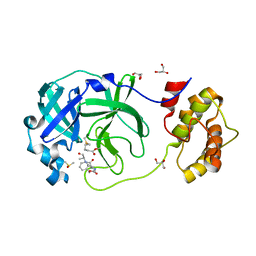 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with Z-VAD-FMK | | 分子名称: | 3C protein, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Lu, M, Yang, H.T, Wang, Z.Y, Zhao, Y, Xing, Y.F. | | 登録日 | 2020-08-24 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Identification of proteasome and caspase inhibitors targeting SARS-CoV-2 M pro .
Signal Transduct Target Ther, 6, 2021
|
|
7CUU
 
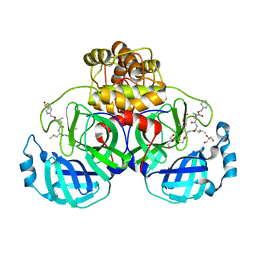 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with MG132 | | 分子名称: | 3C protein, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Lu, M, Yang, H.T, Wang, Z.Y, Zhao, Y, Xing, Y.F. | | 登録日 | 2020-08-24 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Identification of proteasome and caspase inhibitors targeting SARS-CoV-2 M pro .
Signal Transduct Target Ther, 6, 2021
|
|
7E5A
 
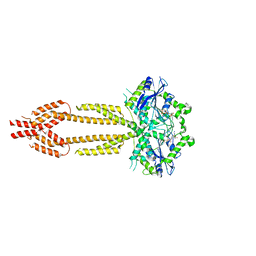 | | interferon-inducible anti-viral protein R356A | | 分子名称: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, Guanylate-binding protein 5, ... | | 著者 | Cui, W, Wang, W, Chen, C, Slater, B, Xiong, Y, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-02-18 | | 公開日 | 2021-05-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7E58
 
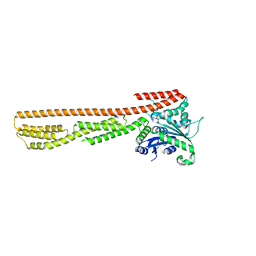 | | interferon-inducible anti-viral protein 2 | | 分子名称: | Guanylate-binding protein 2 | | 著者 | Cui, W, Wang, W, Chen, C, Slater, B, Xiong, Y, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-02-18 | | 公開日 | 2021-05-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7E59
 
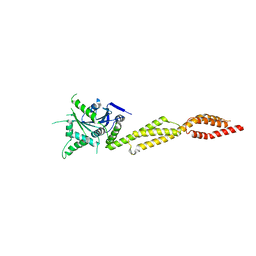 | | interferon-inducible anti-viral protein truncated | | 分子名称: | Guanylate-binding protein 5 | | 著者 | Cui, W, Wang, W, Chen, C, Slater, B, Xiong, Y, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-02-18 | | 公開日 | 2021-05-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3N4M
 
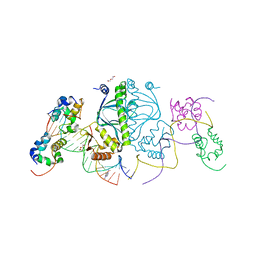 | | E. coli RNA polymerase alpha subunit C-terminal domain in complex with CAP and DNA | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Lara-Gonzalez, S, Birktoft, J.J, Lawson, C.L. | | 登録日 | 2010-05-21 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.987 Å) | | 主引用文献 | The RNA Polymerase alpha Subunit Recognizes the DNA Shape of the Upstream Promoter Element.
Biochemistry, 59, 2020
|
|
7WQV
 
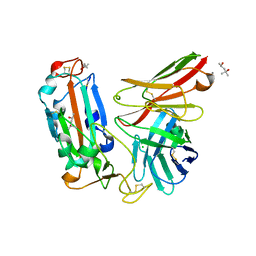 | | Crystal structure of a neutralizing monoclonal antibody (Ab08) in complex with SARS-CoV-2 receptor-binding domain (RBD) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ab08, ... | | 著者 | Zha, J, Meng, L, Zhang, X, Li, D. | | 登録日 | 2022-01-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A Spike-destructing human antibody effectively neutralizes Omicron-included SARS-CoV-2 variants with therapeutic efficacy.
Plos Pathog., 19, 2023
|
|
7EGA
 
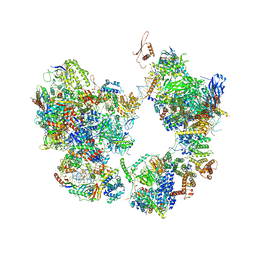 | | TFIID-based intermediate PIC on PUMA promoter | | 分子名称: | DNA (85-MER), DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | 著者 | Chen, X, Wu, Z, Hou, H, Qi, Y, Wang, X, Li, J, Xu, Y. | | 登録日 | 2021-03-24 | | 公開日 | 2021-05-05 | | 最終更新日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EG8
 
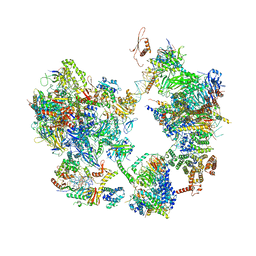 | | TFIID-based core PIC on PUMA promoter | | 分子名称: | DNA (85-MER), DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | 著者 | Chen, X, Qi, Y, Hou, H, Wang, X, Wu, Z, Li, J, Xu, Y. | | 登録日 | 2021-03-24 | | 公開日 | 2021-05-05 | | 最終更新日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (7.4 Å) | | 主引用文献 | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
