7TA0
 
 | | Human Ornithine Aminotransferase (hOAT) soaked with 5-aminovaleric acid | | 分子名称: | 5-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]pentanoic acid, Ornithine aminotransferase, mitochondrial, ... | | 著者 | Butrin, A, Liu, D. | | 登録日 | 2021-12-20 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Determination of the pH dependence, substrate specificity, and turnovers of alternative substrates for human ornithine aminotransferase.
J.Biol.Chem., 298, 2022
|
|
7T9Z
 
 | |
7TA1
 
 | | Human Ornithine Aminotransferase (hOAT) soaked with gamma-Aminobutyric acid | | 分子名称: | 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]BUTANOIC ACID, Ornithine aminotransferase, mitochondrial, ... | | 著者 | Butrin, A, Wawrzak, Z, Liu, D. | | 登録日 | 2021-12-20 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Determination of the pH dependence, substrate specificity, and turnovers of alternative substrates for human ornithine aminotransferase.
J.Biol.Chem., 298, 2022
|
|
6V55
 
 | | Full extracellular region of zebrafish Gpr126/Adgrg6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G-protein coupled receptor G6, CALCIUM ION | | 著者 | Leon, K, Arac, D. | | 登録日 | 2019-12-03 | | 公開日 | 2020-01-15 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Structural basis for adhesion G protein-coupled receptor Gpr126 function.
Nat Commun, 11, 2020
|
|
4PWW
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR494. | | 分子名称: | ACETIC ACID, OR494, PHOSPHATE ION | | 著者 | Vorobiev, S, Lin, Y.-R, Seetharaman, J, Xiao, R, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2014-03-21 | | 公開日 | 2014-04-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.471 Å) | | 主引用文献 | Crystal Structure of Engineered Protein OR494.
To be Published
|
|
3VSF
 
 | | Crystal structure of 1,3Gal43A, an exo-beta-1,3-Galactanase from Clostridium thermocellum | | 分子名称: | GLYCEROL, Ricin B lectin | | 著者 | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | 登録日 | 2012-04-25 | | 公開日 | 2012-12-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.757 Å) | | 主引用文献 | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
6UQ0
 
 | | RNA polymerase II elongation complex with 5-guanidinohydantoin lesion in state 4 | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | 著者 | Oh, J, Wang, D. | | 登録日 | 2019-10-18 | | 公開日 | 2020-06-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.56 Å) | | 主引用文献 | RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3VW7
 
 | | Crystal structure of human protease-activated receptor 1 (PAR1) bound with antagonist vorapaxar at 2.2 angstrom | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Proteinase-activated receptor 1, ... | | 著者 | Zhang, C, Srinivasan, Y, Arlow, D.H, Fung, J.J, Palmer, D, Zheng, Y, Green, H.F, Pandey, A, Dror, R.O, Shaw, D.E, Weis, W.I, Coughlin, S.R, Kobilka, B.K. | | 登録日 | 2012-08-07 | | 公開日 | 2012-12-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | High-resolution crystal structure of human protease-activated receptor 1
Nature, 492, 2012
|
|
6UPX
 
 | | RNA polymerase II elongation complex with 5-guanidinohydantoin lesion in state 1 | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | 著者 | Oh, J, Wang, D. | | 登録日 | 2019-10-18 | | 公開日 | 2020-06-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7TBK
 
 | | Composite structure of the dilated human nuclear pore complex (NPC) symmetric core generated with a 37A in situ cryo-ET map of CD4+ T cell NPC | | 分子名称: | NUP107 CTD, NUP107 NTD, NUP133, ... | | 著者 | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-15 | | 最終更新日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (37 Å) | | 主引用文献 | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7TBI
 
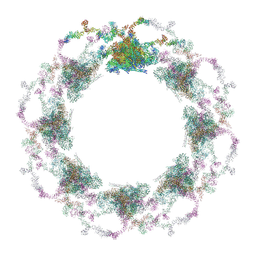 | | Composite structure of the S. cerevisiae nuclear pore complex (NPC) | | 分子名称: | Dyn2, Nic96 R1, Nic96 R2, ... | | 著者 | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-15 | | 最終更新日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (25 Å) | | 主引用文献 | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7TBJ
 
 | | Composite structure of the human nuclear pore complex (NPC) symmetric core generated with a 12A cryo-ET map of the purified HeLa cell NPC | | 分子名称: | NUP107 CTD, NUP107 NTD, NUP133, ... | | 著者 | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-22 | | 最終更新日 | 2022-06-29 | | 実験手法 | ELECTRON MICROSCOPY (23 Å) | | 主引用文献 | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
1WBR
 
 | |
1WCU
 
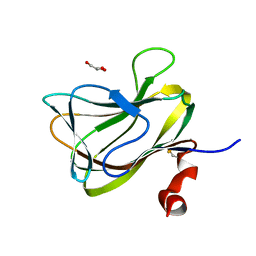 | | CBM29_1, A Family 29 Carbohydrate Binding Module from Piromyces equi | | 分子名称: | GLYCEROL, NON-CATALYTIC PROTEIN 1 | | 著者 | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davis, G.J, Gilbert, H.J. | | 登録日 | 2004-11-22 | | 公開日 | 2005-03-31 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
4Q7C
 
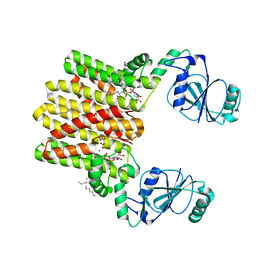 | | Structure of AF2299, a CDP-alcohol phosphotransferase | | 分子名称: | AF2299, a CDP-alcohol phosphotransferase, CALCIUM ION, ... | | 著者 | Clarke, O.B, Sciara, G, Tomasek, D, Banerjee, S, Rajashankar, K.R, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2014-04-24 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.102 Å) | | 主引用文献 | Structural basis for catalysis in a CDP-alcohol phosphotransferase.
Nat Commun, 5, 2014
|
|
3VSZ
 
 | | Crystal structure of Ct1,3Gal43A in complex with galactan | | 分子名称: | GLYCEROL, Ricin B lectin, beta-D-galactopyranose-(1-3)-beta-D-galactopyranose, ... | | 著者 | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | 登録日 | 2012-05-18 | | 公開日 | 2012-12-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.893 Å) | | 主引用文献 | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
6V2N
 
 | | Crystal structure of E. coli phosphoenolpyruvate carboxykinase mutant Lys254Ser | | 分子名称: | ACETATE ION, CALCIUM ION, Phosphoenolpyruvate carboxykinase (ATP) | | 著者 | Sokaribo, A.S, Cotelesage, J.H, Novakovski, B, Goldie, H, Sanders, D. | | 登録日 | 2019-11-25 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Kinetic and structural analysis of Escherichia coli phosphoenolpyruvate carboxykinase mutants.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6UQ2
 
 | | RNA polymerase II elongation complex with dG in state 1 | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | 著者 | Oh, J, Wang, D. | | 登録日 | 2019-10-18 | | 公開日 | 2020-06-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1X99
 
 | | X-ray crystal structure of Xerocomus chrysenteron lectin XCL at 1.4 Angstroms resolution, mutated at Q46M, V54M, L58M | | 分子名称: | 1,2-ETHANEDIOL, SULFATE ION, lectin | | 著者 | Birck, C, Damian, L, Marty-Detraves, C, Lougarre, A, Schulze-Briese, C, Koehl, P, Fournier, D, Paquereau, L, Samama, J.P. | | 登録日 | 2004-08-20 | | 公開日 | 2004-12-14 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | A New Lectin Family with Structure Similarity to Actinoporins Revealed by the Crystal Structure of Xerocomus chrysenteron Lectin XCL
J.Mol.Biol., 344, 2004
|
|
6V67
 
 | | Apo Structure of the De Novo PD-1 Binding Miniprotein GR918.2 | | 分子名称: | PD-1 Binding Miniprotein GR918.2 | | 著者 | Bick, M.J, Bryan, C.M, Baker, D, Dimaio, F, Kang, A. | | 登録日 | 2019-12-04 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.07 Å) | | 主引用文献 | Computational design of a synthetic PD-1 agonist.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7TCI
 
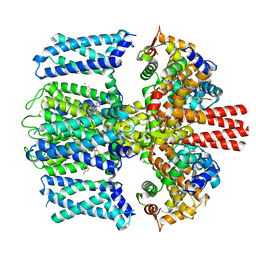 | | Structure of Xenopus KCNQ1-CaM in complex with ML277 | | 分子名称: | (2R)-N-[4-(4-methoxyphenyl)-1,3-thiazol-2-yl]-1-(4-methylbenzene-1-sulfonyl)piperidine-2-carboxamide, CALCIUM ION, Calmodulin-1, ... | | 著者 | Willegems, K, Kyriakis, E, Van Petegem, F, Eldstrom, J, Fedida, D. | | 登録日 | 2021-12-23 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural and electrophysiological basis for the modulation of KCNQ1 channel currents by ML277.
Nat Commun, 13, 2022
|
|
1XBU
 
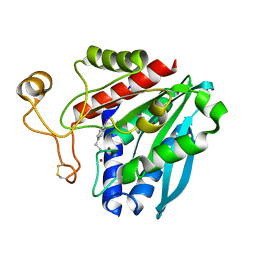 | | Streptomyces griseus aminopeptidase complexed with p-iodo-D-phenylalanine | | 分子名称: | Aminopeptidase, CALCIUM ION, P-IODO-D-PHENYLALANINE, ... | | 著者 | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | 登録日 | 2004-08-31 | | 公開日 | 2005-10-04 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Streptomyces griseus aminopeptidase complexed with p-iodo-D-phenylalanine
To be Published
|
|
7TCP
 
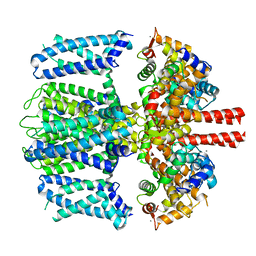 | | Structure of Xenopus KCNQ1-CaM | | 分子名称: | CALCIUM ION, Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 1 | | 著者 | Willegems, K, Kyriakis, E, Van Petegem, F, Eldstrom, J, Fedida, D. | | 登録日 | 2021-12-27 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.84 Å) | | 主引用文献 | Structural and electrophysiological basis for the modulation of KCNQ1 channel currents by ML277.
Nat Commun, 13, 2022
|
|
1XC5
 
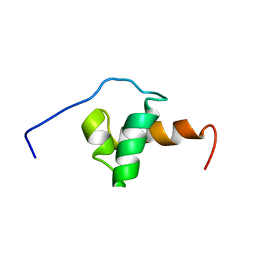 | | Solution Structure of the SMRT Deacetylase Activation Domain | | 分子名称: | Nuclear receptor corepressor 2 | | 著者 | Codina, A, Love, J.D, Li, Y, Lazar, M.A, Neuhaus, D, Schwabe, J.W.R. | | 登録日 | 2004-09-01 | | 公開日 | 2005-05-03 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural insights into the interaction and activation of histone deacetylase 3 by nuclear receptor corepressors
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1XI6
 
 | | Extragenic suppressor from Pyrococcus furiosus Pfu-1862794-001 | | 分子名称: | extragenic suppressor | | 著者 | Zhao, M, Chang, J.C, Zhou, W, Chen, L, Horanyi, P, Xu, H, Yang, H, Liu, Z.-J, Habel, J.E, Lee, D, Chang, S.-H, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2004-09-21 | | 公開日 | 2004-11-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Extragenic suppressor from Pyrococcus furiosus Pfu-1862794-001
To be published
|
|
