7M51
 
 | | B6 Fab fragment bound to the OC43 spike stem helix peptide | | 分子名称: | B6 antigen-binding (Fab) fragment heavy chain, B6 antigen-binding (Fab) fragment light chain, GLYCEROL, ... | | 著者 | Sauer, M.M, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2021-03-22 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for broad coronavirus neutralization.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M55
 
 | | B6 Fab fragment bound to the MERS-CoV spike stem helix peptide | | 分子名称: | B6 antigen binding fragment (Fab) heavy chain, B6 antigen binding fragment (Fab) light chain, GLYCEROL, ... | | 著者 | Sauer, M.M, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2021-03-22 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural basis for broad coronavirus neutralization.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M5E
 
 | |
7M52
 
 | | B6 Fab fragment bound to the HKU4 spike stem helix peptide | | 分子名称: | B6 antigen-binding (Fab) fragment heavy chain, B6 antigen-binding (Fab) fragment light chain, GLYCEROL, ... | | 著者 | Sauer, M.M, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2021-03-22 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural basis for broad coronavirus neutralization.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8HDU
 
 | |
8HDV
 
 | |
7RZU
 
 | |
7RZS
 
 | |
7RZR
 
 | |
7RZT
 
 | |
7RZV
 
 | |
7RZQ
 
 | |
7MCH
 
 | |
2JTC
 
 | | 3D structure and backbone dynamics of SPE B | | 分子名称: | Streptopain | | 著者 | Chuang, W, Wang, C, Houng, H, Chen, C, Wang, P. | | 登録日 | 2007-07-26 | | 公開日 | 2008-08-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and backbone dynamics of streptopain: insight into diverse substrate specificity.
J.Biol.Chem., 284, 2009
|
|
1AUX
 
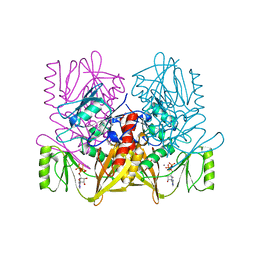 | |
6IR5
 
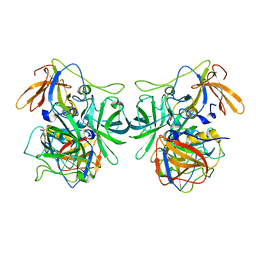 | | P domain of GII.3-TV24 | | 分子名称: | VP1 Capsid protein | | 著者 | Yang, Y. | | 登録日 | 2018-11-10 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses.
Emerg Microbes Infect, 8, 2019
|
|
6IS5
 
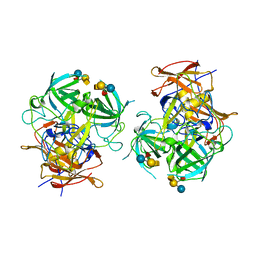 | | P domain of GII.3-TV24 with A-tetrasaccharide complex | | 分子名称: | VP1 Capsid protein, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Yang, Y. | | 登録日 | 2018-11-15 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses.
Emerg Microbes Infect, 8, 2019
|
|
6JBR
 
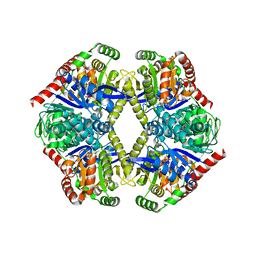 | | Tps1/UDP/T6P complex | | 分子名称: | 6-O-phosphono-alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, Trehalose-6-phosphate synthase, URIDINE-5'-DIPHOSPHATE | | 著者 | Wang, S, Zhao, Y, Wang, D, Liu, J. | | 登録日 | 2019-01-26 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Crystal structures of Magnaporthe oryzae trehalose-6-phosphate synthase (MoTps1) suggest a model for catalytic process of Tps1.
Biochem.J., 476, 2019
|
|
4LNP
 
 | | The first SH3 domain from CAP/Ponsin in complex with proline rich peptide from Vinculin | | 分子名称: | Sorbin and SH3 domain-containing protein 1, Vinculin | | 著者 | Zhao, D, Li, F, Wu, J, Shi, Y, Zhang, Z, Gong, Q. | | 登録日 | 2013-07-11 | | 公開日 | 2014-05-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Structural investigation of the interaction between the tandem SH3 domains of c-Cbl-associated protein and vinculin
J.Struct.Biol., 187, 2014
|
|
7WPO
 
 | | Structure of NeoCOV RBD binding to Bat37 ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | 登録日 | 2022-01-24 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
7WPZ
 
 | | Structure of PDF-2180-COV RBD binding to Bat37 ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | 著者 | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | 登録日 | 2022-01-24 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
7JOU
 
 | |
1KX9
 
 | | ANTENNAL CHEMOSENSORY PROTEIN A6 FROM THE MOTH MAMESTRA BRASSICAE | | 分子名称: | ACETATE ION, CHEMOSENSORY PROTEIN A6 | | 著者 | Lartigue, A, Campanacci, V, Roussel, A, Larsson, A.M, Jones, T.A, Tegoni, M, Cambillau, C. | | 登録日 | 2002-01-31 | | 公開日 | 2002-12-04 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | X-ray structure and ligand binding study of a moth chemosensory protein
J.Biol.Chem., 277, 2002
|
|
4LNU
 
 | | Nucleotide-free kinesin motor domain in complex with tubulin and a DARPin | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Designed ankyrin repeat protein (DARPIN) D1, GLYCEROL, ... | | 著者 | Cao, L, Gigant, B, Knossow, M. | | 登録日 | 2013-07-12 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | The structure of apo-kinesin bound to tubulin links the nucleotide cycle to movement
Nat Commun, 5, 2014
|
|
1NAW
 
 | | ENOLPYRUVYL TRANSFERASE | | 分子名称: | CYCLOHEXYLAMMONIUM ION, UDP-N-ACETYLGLUCOSAMINE 1-CARBOXYVINYL-TRANSFERASE | | 著者 | Schoenbrunn, E, Sack, S, Eschenburg, S, Perrakis, A, Krekel, F, Amrhein, N, Mandelkow, E. | | 登録日 | 1996-07-23 | | 公開日 | 1997-07-23 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of UDP-N-acetylglucosamine enolpyruvyltransferase, the target of the antibiotic fosfomycin.
Structure, 4, 1996
|
|
