1HJP
 
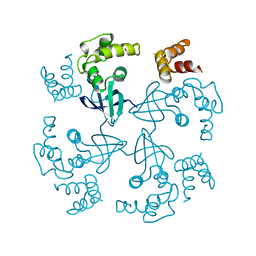 | | HOLLIDAY JUNCTION BINDING PROTEIN RUVA FROM E. COLI | | 分子名称: | RUVA | | 著者 | Nishino, T, Ariyoshi, M, Iwasaki, H, Shinagawa, H, Morikawa, K. | | 登録日 | 1997-08-21 | | 公開日 | 1998-02-25 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Functional Analyses of the Domain Structure in the Holliday Junction Binding Protein Ruva
Structure, 6, 1998
|
|
1IUG
 
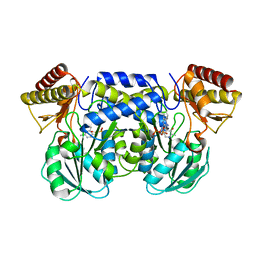 | | The crystal structure of aspartate aminotransferase which belongs to subgroup IV from Thermus thermophilus | | 分子名称: | PHOSPHATE ION, putative aspartate aminotransferase | | 著者 | Katsura, Y, Shirouzu, M, Yamaguchi, H, Ishitani, R, Nureki, O, Kuramitsu, S, Hayashi, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2002-03-04 | | 公開日 | 2003-11-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of a putative aspartate aminotransferase belonging to subgroup IV.
Proteins, 55, 2004
|
|
4EN6
 
 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-3-sialyllactose | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | 登録日 | 2012-04-12 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
4EN7
 
 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-3-sialyllactosamine | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | 登録日 | 2012-04-12 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
4EN8
 
 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-6-sialyllactose | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | 登録日 | 2012-04-12 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
1IXS
 
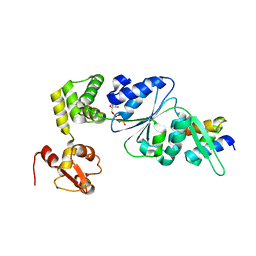 | | Structure of RuvB complexed with RuvA domain III | | 分子名称: | Holliday junction DNA helicase ruvA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RuvB | | 著者 | Yamada, K, Miyata, T, Tsuchiya, D, Oyama, T, Fujiwara, Y, Ohnishi, T, Iwasaki, H, Shinagawa, H, Ariyoshi, M, Mayanagi, K, Morikawa, K. | | 登録日 | 2002-07-04 | | 公開日 | 2002-11-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal Structure of the RuvA-RuvB Complex: A Structural Basis for the Holliday Junction Migrating Motor Machinery
Mol.Cell, 10, 2002
|
|
1J0S
 
 | | Solution structure of the human interleukin-18 | | 分子名称: | Interleukin-18 | | 著者 | Kato, Z, Jee, J, Shikano, H, Mishima, M, Ohki, I, Yoneda, T, Hara, T, Torigoe, K, Kondo, N, Shirakawa, M. | | 登録日 | 2002-11-21 | | 公開日 | 2003-11-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structure and binding mode of interleukin-18
Nat.Struct.Biol., 10, 2003
|
|
1IXR
 
 | | RuvA-RuvB complex | | 分子名称: | Holliday junction DNA helicase ruvA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RuvB | | 著者 | Yamada, K, Miyata, T, Tsuchiya, D, Oyama, T, Fujiwara, Y, Ohnishi, T, Iwasaki, H, Shinagawa, H, Ariyoshi, M, Mayanagi, K, Morikawa, K. | | 登録日 | 2002-07-04 | | 公開日 | 2002-11-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Crystal Structure of the RuvA-RuvB Complex: A Structural Basis for the Holliday Junction Migrating Motor Machinery
Mol.Cell, 10, 2002
|
|
1ZBY
 
 | | High-Resolution Crystal Structure of Native (Resting) Cytochrome c Peroxidase (CcP) | | 分子名称: | Cytochrome c peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Bonagura, C.A, Bhaskar, B, Shimizu, H, Li, H, Sundaramoorthy, M, McRee, D.E, Goodin, D.B, Poulos, T.L. | | 登録日 | 2005-04-09 | | 公開日 | 2005-05-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | High-resolution crystal structures and spectroscopy of native and compound I cytochrome c peroxidase
Biochemistry, 42, 2003
|
|
7V5N
 
 | | Crystal structure of Fab fragment of bevacizumab bound to DNA aptamer | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*GP*TP*TP*GP*GP*TP*GP*GP*TP*AP*GP*TP*TP*AP*CP*GP*TP*TP*CP*GP*C)-3'), IMIDAZOLE, ... | | 著者 | Hishiki, A, Tong, J, Todoroki, K, Hashimoto, H. | | 登録日 | 2021-08-17 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Development of a DNA aptamer that binds to the complementarity-determining region of therapeutic monoclonal antibody and affinity improvement induced by pH-change for sensitive detection.
Biosens.Bioelectron., 203, 2022
|
|
8GNN
 
 | | Crystal structure of the human RAD9-RAD1-HUS1-RAD17 complex | | 分子名称: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Cell cycle checkpoint protein RAD17, ... | | 著者 | Hara, K, Nagata, K, Iida, N, Hashimoto, H. | | 登録日 | 2022-08-24 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.119 Å) | | 主引用文献 | The 9-1-1 DNA clamp subunit RAD1 forms specific interactions with clamp loader RAD17, revealing functional implications for binding-protein RHINO.
J.Biol.Chem., 299, 2023
|
|
1KOZ
 
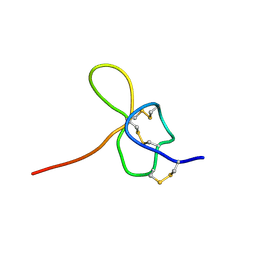 | | SOLUTION STRUCTURE OF OMEGA-GRAMMOTOXIN SIA | | 分子名称: | Voltage-dependent Channel Inhibitor | | 著者 | Takeuchi, K, Park, E.J, Lee, C.W, Kim, J.I, Takahashi, H, Swartz, K.J, Shimada, I. | | 登録日 | 2001-12-25 | | 公開日 | 2002-08-28 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of omega-grammotoxin SIA, a gating modifier of P/Q and N-type Ca(2+) channel.
J.Mol.Biol., 321, 2002
|
|
7DEN
 
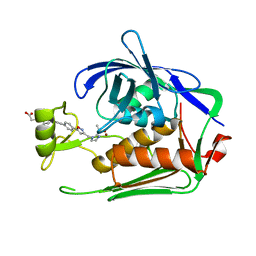 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | 分子名称: | 4-[(1~{R},5~{S})-6-[2-[4-[3-[[2-[(1~{S})-1-oxidanylethyl]imidazol-1-yl]methyl]-1,2-oxazol-5-yl]phenyl]ethynyl]-3-azabicyclo[3.1.0]hexan-3-yl]butanoic acid, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | 著者 | Mima, M, Ushiyama, F, Takashima, H. | | 登録日 | 2020-11-04 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Lead optimization of 2-hydroxymethyl imidazoles as non-hydroxamate LpxC inhibitors: Discovery of TP0586532.
Bioorg.Med.Chem., 30, 2020
|
|
7XHT
 
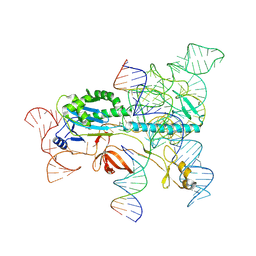 | | Structure of the OgeuIscB-omega RNA-target DNA complex | | 分子名称: | DNA (49-MER), DNA (5'-D(P*GP*AP*AP*GP*AP*AP*AP*AP*CP*CP*AP*T)-3'), LAURYL DIMETHYLAMINE-N-OXIDE, ... | | 著者 | Kato, K, Okazaki, O, Isayama, Y, Ishikawa, J, Nishizawa, T, Nishimasu, H. | | 登録日 | 2022-04-10 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.55 Å) | | 主引用文献 | Structure of the IscB-omega RNA ribonucleoprotein complex, the likely ancestor of CRISPR-Cas9.
Nat Commun, 13, 2022
|
|
1ZBZ
 
 | | High-Resolution Crystal Structure of Compound I intermediate of Cytochrome c Peroxidase (CcP) | | 分子名称: | Cytochrome c peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Bonagura, C.A, Bhaskar, B, Shimizu, H, Li, H, Sundaramoorthy, M, McRee, D.E, Goodin, D.B, Poulos, T.L. | | 登録日 | 2005-04-09 | | 公開日 | 2005-05-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | High-resolution crystal structures and spectroscopy of native and compound I cytochrome c peroxidase
Biochemistry, 42, 2003
|
|
1DLF
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE FV FRAGMENT FROM AN ANTI-DANSYL SWITCH VARIANT ANTIBODY IGG2A(S) CRYSTALLIZED AT PH 5.25 | | 分子名称: | ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S), SULFATE ION | | 著者 | Nakasako, M, Takahashi, H, Shimada, I, Arata, Y. | | 登録日 | 1998-07-14 | | 公開日 | 1999-07-26 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the Fv fragment from an anti-dansyl monoclonal antibody.
J.Mol.Biol., 291, 1999
|
|
5YKY
 
 | | Crystal Structure of Cross-Linked Tetragonal Hen Egg White Lysozyme Soaked with 10 mM Rose Bengal and 10mM H2PtCl6 | | 分子名称: | CHLORIDE ION, Lysozyme C, PLATINUM (II) ION, ... | | 著者 | Tabe, H, Takahashi, H, Shimoi, T, Abe, S, Ueno, T, Yamada, Y. | | 登録日 | 2017-10-16 | | 公開日 | 2018-09-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Photocatalytic hydrogen evolution systems constructed in cross-linked porous protein crystals
Appl Catal B, 237, 2018
|
|
3AAM
 
 | | Crystal structure of endonuclease IV from Thermus thermophilus HB8 | | 分子名称: | Endonuclease IV, MANGANESE (II) ION, PHOSPHATE ION | | 著者 | Asano, R, Ishikawa, H, Nakane, S, Baba, S, Nakagawa, N, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-11-20 | | 公開日 | 2010-12-01 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | An additional C-terminal loop in endonuclease IV, an apurinic/apyrimidinic endonuclease, controls binding affinity to DNA
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3A9R
 
 | | X-ray Structures of Bacillus pallidus D-Arabinose IsomeraseComplex with (4R)-2-METHYLPENTANE-2,4-DIOL | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, D-arabinose isomerase, MANGANESE (II) ION | | 著者 | Takeda, K, Yoshida, H, Izumori, K, Kamitori, S. | | 登録日 | 2009-11-05 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | X-ray structures of Bacillus pallidusd-arabinose isomerase and its complex with l-fucitol.
Biochim.Biophys.Acta, 1804, 2010
|
|
3AEX
 
 | |
3AAL
 
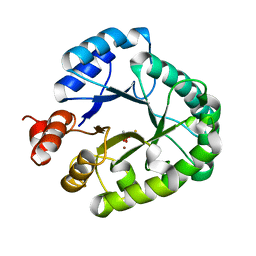 | | Crystal Structure of endonuclease IV from Geobacillus kaustophilus | | 分子名称: | CACODYLATE ION, FE (III) ION, Probable endonuclease 4, ... | | 著者 | Asano, R, Ishikawa, H, Nakane, S, Nakagawa, N, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-11-20 | | 公開日 | 2010-12-01 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | An additional C-terminal loop in endonuclease IV, an apurinic/apyrimidinic endonuclease, controls binding affinity to DNA
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3A9T
 
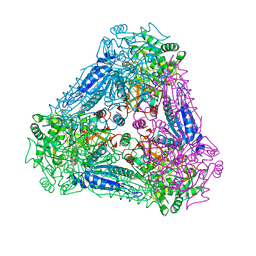 | | X-ray Structure of Bacillus pallidus D-Arabinose Isomerase Complex with L-Fucitol | | 分子名称: | D-arabinose isomerase, FUCITOL, MANGANESE (II) ION | | 著者 | Takeda, K, Yoshida, H, Izumori, K, Kamitori, S. | | 登録日 | 2009-11-05 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | X-ray structures of Bacillus pallidusd-arabinose isomerase and its complex with l-fucitol.
Biochim.Biophys.Acta, 1804, 2010
|
|
3AK9
 
 | | Crystal structure of the SEp22 dodecamer, a Dps-like protein from Salmonella enterica subsp. enterica serovar Enteritidis, FE-soaked form | | 分子名称: | DNA protection during starvation protein, FE (II) ION, MAGNESIUM ION, ... | | 著者 | Miyamoto, T, Asahina, Y, Miyazaki, S, Shimizu, H, Ohto, U, Noguchi, S, Satow, Y. | | 登録日 | 2010-07-08 | | 公開日 | 2011-01-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structures of the SEp22 dodecamer, a Dps-like protein from Salmonella enterica subsp. enterica serovar Enteritidis
Acta Crystallogr.,Sect.F, 67, 2011
|
|
6JP6
 
 | | The X-ray structure of yeast tRNA methyltransferase complex of Trm7 and Trm734 essential for 2'-O-methylation at the first position of anticodon in specific tRNAs | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, tRNA (cytidine(34)/guanosine(34)-2'-O)-methyltransferase, ... | | 著者 | Hirata, A, Okada, K, Yoshii, K, Shiraisi, H, Saijo, S, Yonezawa, K, Sihimzu, N, Hori, H. | | 登録日 | 2019-03-26 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.699 Å) | | 主引用文献 | Structure of tRNA methyltransferase complex of Trm7 and Trm734 reveals a novel binding interface for tRNA recognition.
Nucleic Acids Res., 47, 2019
|
|
7C7L
 
 | | Cryo-EM structure of the Cas12f1-sgRNA-target DNA complex | | 分子名称: | CRISPR-associated protein Cas14a.1, DNA (40-mer), ZINC ION, ... | | 著者 | Takeda, N.S, Nakagawa, R, Okazaki, S, Hirano, H, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | 登録日 | 2020-05-26 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of the miniature type V-F CRISPR-Cas effector enzyme.
Mol.Cell, 81, 2021
|
|
