3NI6
 
 | |
5CYN
 
 | |
7KEF
 
 | | RNA polymerase II elongation complex with unnatural base dTPT3, rNaM in swing state | | 分子名称: | (1S)-1,4-anhydro-1-(3-methoxynaphthalen-2-yl)-5-O-phosphono-D-ribitol, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | 著者 | Oh, J, Wang, D. | | 登録日 | 2020-10-10 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.89 Å) | | 主引用文献 | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
7KED
 
 | | RNA polymerase II elongation complex with unnatural base dTPT3 | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | 著者 | Oh, J, Wang, W, Wang, D. | | 登録日 | 2020-10-10 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
7KEE
 
 | | RNA polymerase II elongation complex with unnatural base dTPT3, rNaMTP bound to E-site | | 分子名称: | (1S)-1,4-anhydro-5-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-1-(3-methoxynaphthalen-2-yl)-D-ribitol, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | 著者 | Oh, J, Wang, D. | | 登録日 | 2020-10-10 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
4NBP
 
 | |
3IHZ
 
 | | Crystal structure of the FK506 binding domain of Plasmodium vivax FKBP35 in complex with FK506 | | 分子名称: | 70 kDa peptidylprolyl isomerase, putative, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN | | 著者 | Qureshi, I.A, Alag, R, Yoon, H.S, Lescar, J. | | 登録日 | 2009-07-31 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | NMR and crystallographic structures of the FK506 binding domain of human malarial parasite Plasmodium vivax FKBP35
Protein Sci., 19, 2010
|
|
2HQR
 
 | |
2HQO
 
 | |
2HQN
 
 | |
4ITZ
 
 | |
4LIF
 
 | |
4LMD
 
 | |
6L89
 
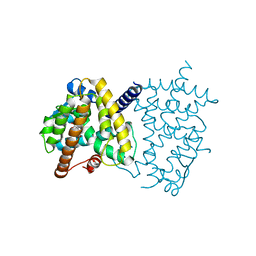 | | Human PPARgamma ligand binding domain complexed with Butyrolactone 1 | | 分子名称: | Peroxisome proliferator-activated receptor gamma, methyl (2R)-3-(4-hydroxyphenyl)-2-[[3-(3-methylbut-2-enyl)-4-oxidanyl-phenyl]methyl]-4-oxidanyl-5-oxidanylidene-furan-2-carboxylate | | 著者 | Jang, D.M, Han, B.W. | | 登録日 | 2019-11-05 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Cyclin-Dependent Kinase 5 Inhibitor Butyrolactone I Elicits a Partial Agonist Activity of Peroxisome Proliferator-Activated Receptor gamma.
Biomolecules, 10, 2020
|
|
6L8B
 
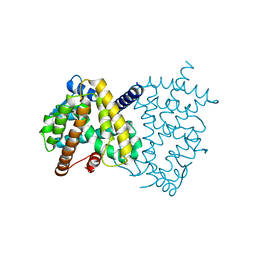 | | The ligand-free structure of human PPARgamma LBD | | 分子名称: | Peroxisome proliferator-activated receptor gamma | | 著者 | Jang, D.M, Han, B.W. | | 登録日 | 2019-11-05 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.102 Å) | | 主引用文献 | Cyclin-Dependent Kinase 5 Inhibitor Butyrolactone I Elicits a Partial Agonist Activity of Peroxisome Proliferator-Activated Receptor gamma.
Biomolecules, 10, 2020
|
|
1OGF
 
 | |
1OGE
 
 | |
1OGC
 
 | |
1OGD
 
 | |
1OHU
 
 | |
1IQS
 
 | | Minimized average structure of MTH1880 from Methanobacterium Thermoautotrophicum | | 分子名称: | MTH1880 | | 著者 | Lee, C.H, Shin, J, Bang, E, Jung, J.W, Yee, A, Arrowsmith, C.H, Lee, W. | | 登録日 | 2001-07-29 | | 公開日 | 2002-07-29 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a novel calcium binding protein, MTH1880, from Methanobacterium thermoautotrophicum.
Protein Sci., 13, 2004
|
|
1IQO
 
 | | Solution structure of MTH1880 from methanobacterium thermoautotrophicum | | 分子名称: | HYPOTHETICAL PROTEIN MTH1880 | | 著者 | Lee, C.H, Shin, J, Bang, E, Jung, J.W, Yee, A, Arrowsmith, C.H, Lee, W. | | 登録日 | 2001-07-23 | | 公開日 | 2002-07-24 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a novel calcium binding protein, MTH1880, from Methanobacterium thermoautotrophicum.
Protein Sci., 13, 2004
|
|
6M1B
 
 | | A new V27M variant of beta 2 microglobulin induced amyloidosis in a patient with long-term hemodialysis | | 分子名称: | Beta-2-microglobulin, CALCIUM ION, GLYCEROL, ... | | 著者 | So, M, Nakahara, S, Nakaniwa, T, Tanaka, H, Kurisu, G, Goto, Y. | | 登録日 | 2020-02-25 | | 公開日 | 2021-01-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Dialysis-related amyloidosis associated with a novel beta 2 -microglobulin variant.
Amyloid, 28, 2021
|
|
