3L1Y
 
 | |
6CO1
 
 | |
6D0L
 
 | | Structure of human TIRR | | 分子名称: | Tudor-interacting repair regulator protein | | 著者 | Cui, G, Botuyan, M.V, Mer, G. | | 登録日 | 2018-04-10 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5VF0
 
 | |
6VE5
 
 | |
5VZM
 
 | |
7LYC
 
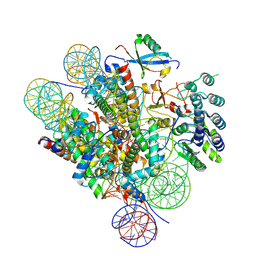 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A Lys13 and Lys15 in complex with BARD1 (residues 415-777) | | 分子名称: | BRCA1-associated RING domain protein 1, DNA (146-MER), DNA (147-MER), ... | | 著者 | Hu, Q, Botuyan, M.V, Zhao, D, Cui, D, Mer, E, Mer, G. | | 登録日 | 2021-03-06 | | 公開日 | 2021-06-16 | | 最終更新日 | 2021-09-01 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | Mechanisms of BRCA1-BARD1 nucleosome recognition and ubiquitylation.
Nature, 596, 2021
|
|
7LYA
 
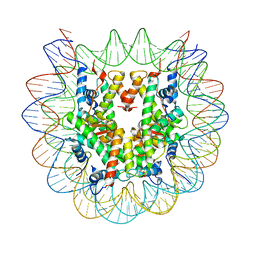 | | Cryo-EM structure of the human nucleosome core particle with linked histone proteins H2A and H2B | | 分子名称: | DNA (146-MER), DNA (147-MER), Histone H2A type 1-B/E, ... | | 著者 | Hu, Q, Botuyan, M.V, Zhao, D, Cui, D, Mer, E, Mer, G. | | 登録日 | 2021-03-06 | | 公開日 | 2021-07-28 | | 最終更新日 | 2021-09-01 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | Mechanisms of BRCA1-BARD1 nucleosome recognition and ubiquitylation.
Nature, 596, 2021
|
|
7LYB
 
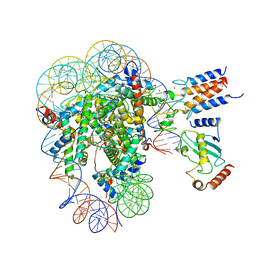 | | Cryo-EM structure of the human nucleosome core particle in complex with BRCA1-BARD1-UbcH5c | | 分子名称: | BRCA1-associated RING domain protein 1, DNA (146-MER), DNA (147-MER), ... | | 著者 | Hu, Q, Botuyan, M.V, Zhao, D, Cui, D, Mer, E, Mer, G. | | 登録日 | 2021-03-06 | | 公開日 | 2021-07-28 | | 最終更新日 | 2021-09-01 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Mechanisms of BRCA1-BARD1 nucleosome recognition and ubiquitylation.
Nature, 596, 2021
|
|
3P8D
 
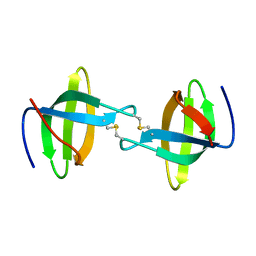 | | Crystal structure of the second Tudor domain of human PHF20 (homodimer form) | | 分子名称: | Medulloblastoma antigen MU-MB-50.72 | | 著者 | Cui, G, Lee, J, Thompson, J.R, Botuyan, M.V, Mer, G. | | 登録日 | 2010-10-13 | | 公開日 | 2011-06-22 | | 最終更新日 | 2012-09-26 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | PHF20 is an effector protein of p53 double lysine methylation that stabilizes and activates p53.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3PD7
 
 | |
3FSS
 
 | |
5VWE
 
 | |
2K2W
 
 | | Second BRCT domain of NBS1 | | 分子名称: | Recombination and DNA repair protein | | 著者 | Xu, C, Cui, G, Botuyan, M, Mer, G. | | 登録日 | 2008-04-14 | | 公開日 | 2008-06-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of a second BRCT domain identified in the nijmegen breakage syndrome protein Nbs1 and its function in an MDC1-dependent localization of Nbs1 to DNA damage sites.
J.Mol.Biol., 381, 2008
|
|
2KTF
 
 | |
3PA6
 
 | |
2L0F
 
 | |
2L0G
 
 | |
2LH9
 
 | |
2LDM
 
 | |
2LH0
 
 | |
2QQS
 
 | |
2LVM
 
 | |
2LY1
 
 | |
2LY2
 
 | |
