7YHL
 
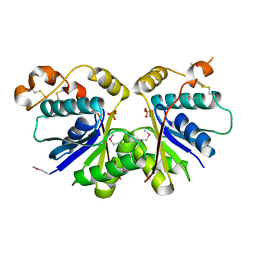 | |
7YGH
 
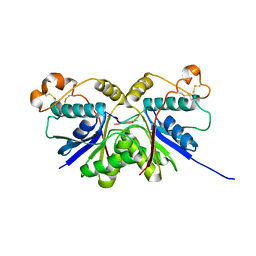 | |
5Z6Q
 
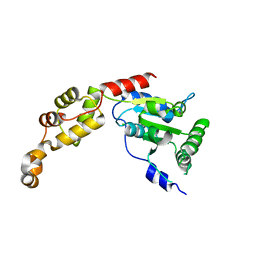 | |
5ZGG
 
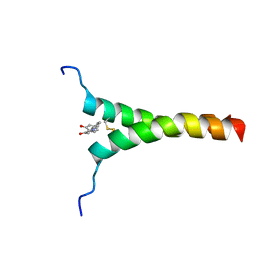 | |
3UB2
 
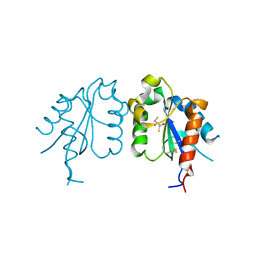 | | TIR domain of Mal/TIRAP | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Toll/interleukin-1 receptor domain-containing adapter protein | | 著者 | Shen, Y, Lin, Z. | | 登録日 | 2011-10-23 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Insights into TIR Domain Specificity of the Bridging Adaptor Mal in TLR4 Signaling
Plos One, 7, 2012
|
|
7DFE
 
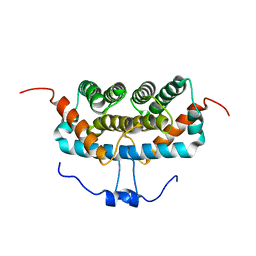 | | NMR structure of TuSp2-RP | | 分子名称: | B6 protein | | 著者 | Lin, Z, Fan, T, Fan, J. | | 登録日 | 2020-11-07 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | 1H, 15N and 13C resonance assignments of a repetitive domain of tubuliform spidroin 2
Biomol.Nmr Assign., 15, 2021
|
|
7CSQ
 
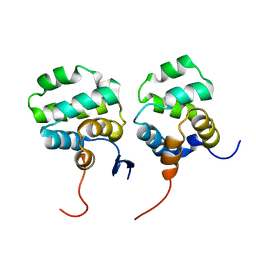 | | Solution structure of the complex between p75NTR-DD and TRADD-DD | | 分子名称: | Tumor necrosis factor receptor superfamily member 16, Tumor necrosis factor receptor type 1-associated DEATH domain protein | | 著者 | Lin, Z, Zhang, N. | | 登録日 | 2020-08-16 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis of NF-kappa B signaling by the p75 neurotrophin receptor interaction with adaptor protein TRADD through their respective death domains.
J.Biol.Chem., 297, 2021
|
|
7X5B
 
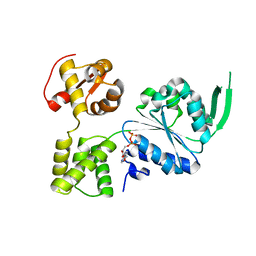 | | Crystal structure of RuvB | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvB | | 著者 | Lin, Z, Qu, Q, Zhang, X, Zhou, Z, Dai, L. | | 登録日 | 2022-03-04 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X5A
 
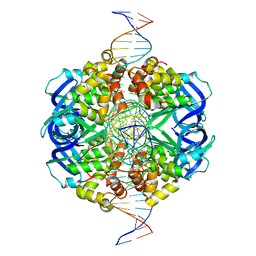 | | CryoEM structure of RuvA-Holliday junction complex | | 分子名称: | DNA (26-MER), Holliday junction ATP-dependent DNA helicase RuvA | | 著者 | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | 登録日 | 2022-03-04 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X7P
 
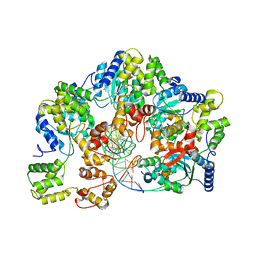 | | CryoEM structure of dsDNA-RuvB-RuvA domain3 complex | | 分子名称: | DNA, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB | | 著者 | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | 登録日 | 2022-03-10 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (7.02 Å) | | 主引用文献 | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X7Q
 
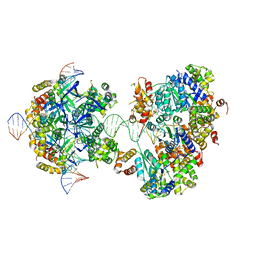 | | CryoEM structure of RuvA-RuvB-Holliday junction complex | | 分子名称: | DNA (26-MER), DNA (40-MER), Holliday junction ATP-dependent DNA helicase RuvA, ... | | 著者 | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | 登録日 | 2022-03-10 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (7.02 Å) | | 主引用文献 | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
4HRV
 
 | |
2VTF
 
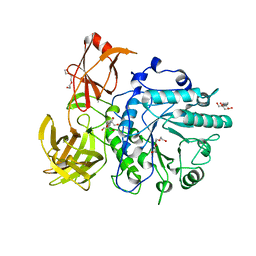 | | X-ray crystal structure of the Endo-beta-N-acetylglucosaminidase from Arthrobacter protophormiae E173Q mutant reveals a TIM barrel catalytic domain and two ancillary domains | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ENDO-BETA-N-ACETYLGLUCOSAMINIDASE, TRIETHYLENE GLYCOL | | 著者 | Ling, Z, Bingham, R.J, Suits, M.D.L, Moir, J.W.B, Fairbanks, A.J, Taylor, E.J. | | 登録日 | 2008-05-14 | | 公開日 | 2009-03-31 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | The X-Ray Crystal Structure of an Arthrobacter Protophormiae Endo-Beta-N-Acetylglucosaminidase Reveals a (Beta/Alpha)(8) Catalytic Domain, Two Ancillary Domains and Active Site Residues Key for Transglycosylation Activity.
J.Mol.Biol., 389, 2009
|
|
6LW3
 
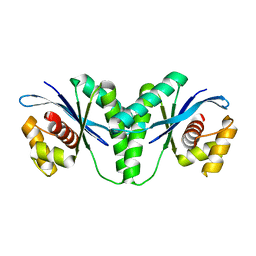 | | Crystal structure of RuvC from Pseudomonas aeruginosa | | 分子名称: | Crossover junction endodeoxyribonuclease RuvC | | 著者 | Hu, Y, He, Y, Lin, Z. | | 登録日 | 2020-02-07 | | 公開日 | 2020-02-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Biochemical and structural characterization of the Holliday junction resolvase RuvC from Pseudomonas aeruginosa.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
8HQ2
 
 | |
4XSK
 
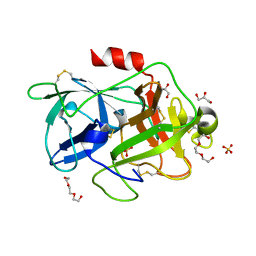 | | Structure of PAItrap, an uPA mutant | | 分子名称: | GLYCEROL, SULFATE ION, TRIETHYLENE GLYCOL, ... | | 著者 | Gong, L, Proulle, V, Hong, Z, Lin, Z, Liu, M, Yuan, C, Lin, L, Furie, B, Flaumenhaft, R, Andreasen, P, Furie, B, Huang, M. | | 登録日 | 2015-01-22 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure of PAItrap, an uPA mutant
To Be Published
|
|
4XKL
 
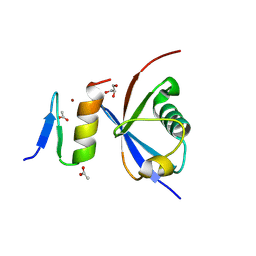 | | Crystal structure of NDP52 ZF2 in complex with mono-ubiquitin | | 分子名称: | ACETATE ION, Calcium-binding and coiled-coil domain-containing protein 2, GLYCEROL, ... | | 著者 | Xie, X, Li, F, Wang, Y, Lin, Z, Chen, X, Liu, J, Pan, L. | | 登録日 | 2015-01-12 | | 公開日 | 2015-11-11 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Molecular basis of ubiquitin recognition by the autophagy receptor CALCOCO2
Autophagy, 11, 2015
|
|
6HCF
 
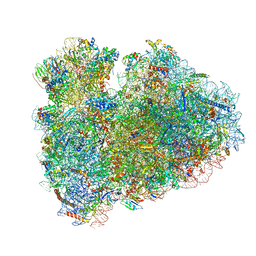 | | Structure of the rabbit 80S ribosome stalled on globin mRNA at the stop codon | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | 著者 | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | 登録日 | 2018-08-14 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
8GUG
 
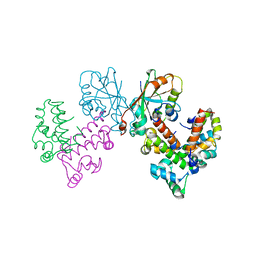 | | Structure of VPA0770 toxin bound to VPA0769 antitoxin in Vibrio parahaemolyticus | | 分子名称: | DUF2384 domain-containing protein, RES domain-containing protein | | 著者 | Song, X.J, Zhang, Y, Xu, Y.Y, Lin, Z. | | 登録日 | 2022-09-12 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural insights of the toxin-antitoxin system VPA0770-VPA0769 in Vibrio parahaemolyticus.
Int.J.Biol.Macromol., 242, 2023
|
|
1M8S
 
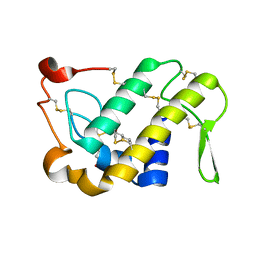 | | Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 5.9) | | 分子名称: | 1,4-BUTANEDIOL, CADMIUM ION, phospholipase a2 | | 著者 | Xu, S, Gu, L, Zhou, Y, Lin, Z. | | 登録日 | 2002-07-25 | | 公開日 | 2003-02-11 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures of cadmium-binding acidic phospholipase A(2) from the venom of Agkistrodon halys Pallas at 1.9A resolutio
Biochem.Biophys.Res.Commun., 300, 2003
|
|
1M8R
 
 | | Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 7.4) | | 分子名称: | 1,4-BUTANEDIOL, CADMIUM ION, phospholipase A2 | | 著者 | Xu, S, Gu, L, Zhou, Y, Lin, Z. | | 登録日 | 2002-07-25 | | 公開日 | 2003-02-11 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures of cadmium-binding acidic phospholipase A(2) from the venom of Agkistrodon halys Pallas at 1.9A resolutio
Biochem.Biophys.Res.Commun., 300, 2003
|
|
5V6J
 
 | | Glycan binding protein Y3 from mushroom Coprinus comatus possesses anti-leukemic activity | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, TMV resistance protein Y3 | | 著者 | Li, K, Zhang, P, Gang, Y, Xia, C, Polston, J.E, Li, G, Li, S, Lin, Z, Yang, L.-J, Bruner, S.D, Ding, Y. | | 登録日 | 2017-03-16 | | 公開日 | 2017-08-16 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.18 Å) | | 主引用文献 | Cytotoxic protein from the mushroom Coprinus comatus possesses a unique mode for glycan binding and specificity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8KFT
 
 | | Crystal structure of ZmMOC1 in complex with a nicked Holliday junction soaked in Mn2+ for 15 seconds | | 分子名称: | DNA (25-MER), DNA (33-MER), DNA (5'-D(P*CP*AP*CP*GP*AP*TP*TP*G)-3'), ... | | 著者 | Zhang, D, Luo, Z, Lin, Z. | | 登録日 | 2023-08-16 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFV
 
 | | Crystal structure of ZmMOC1 K229A in complex with a nicked Holliday junction soaked in Mn2+ for 180 seconds | | 分子名称: | 1,2-ETHANEDIOL, DNA (25-MER), DNA (33-MER), ... | | 著者 | Zhang, D, Luo, Z, Lin, Z. | | 登録日 | 2023-08-16 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFU
 
 | | Crystal structure of ZmMOC1 in complex with a nicked Holliday junction soaked in Mn2+ for 180 seconds | | 分子名称: | 1,2-ETHANEDIOL, DNA (25-MER), DNA (33-MER), ... | | 著者 | Zhang, D, Luo, Z, Lin, Z. | | 登録日 | 2023-08-16 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
