8X15
 
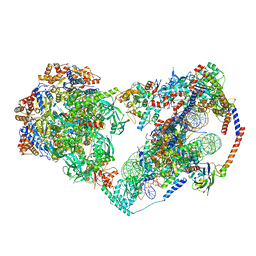 | | Structure of nucleosome-bound SRCAP-C in the apo state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | 著者 | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | 登録日 | 2023-11-06 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
8X19
 
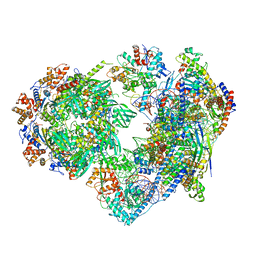 | | Structure of nucleosome-bound SRCAP-C in the ADP-BeFx-bound state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | 著者 | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | 登録日 | 2023-11-06 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
6VAJ
 
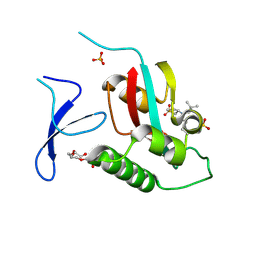 | | Crystal Structure Analysis of human PIN1 | | 分子名称: | 2-chloro-N-(2,2-dimethylpropyl)-N-[(3R)-1,1-dioxo-1lambda~6~-thiolan-3-yl]acetamide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION, ... | | 著者 | Seo, H.-S, Dhe-Paganon, S. | | 登録日 | 2019-12-17 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Sulfopin is a covalent inhibitor of Pin1 that blocks Myc-driven tumors in vivo.
Nat.Chem.Biol., 17, 2021
|
|
8XS5
 
 | |
8XNG
 
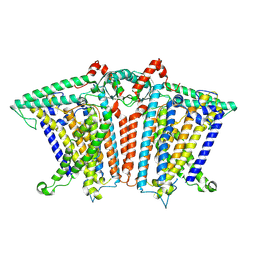 | |
8XW4
 
 | |
8XW0
 
 | | Cryo-EM structure of OSCA3.1-GDN state | | 分子名称: | CSC1-like protein ERD4, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine | | 著者 | Zhang, Y, Han, Y. | | 登録日 | 2024-01-15 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XW3
 
 | |
8XRY
 
 | |
8XW2
 
 | |
8XVZ
 
 | |
8XAJ
 
 | |
8XW1
 
 | |
8XVX
 
 | |
8XS0
 
 | |
8XS4
 
 | |
8XVY
 
 | |
6M3M
 
 | |
6LNQ
 
 | | The co-crystal structure of SARS-CoV 3C Like Protease with aldehyde inhibitor M7 | | 分子名称: | N-[(2S)-3-methyl-1-[[(2S)-4-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]-1H-indole-2-carboxamide, Severe Acute Respiratory Syndrome Coronavirus 3c Like Protease | | 著者 | Wang, H, Shang, L.Q. | | 登録日 | 2020-01-01 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.244 Å) | | 主引用文献 | Comprehensive Insights into the Catalytic Mechanism of Middle East Respiratory Syndrome 3C-Like Protease and Severe Acute Respiratory Syndrome 3C-Like Protease.
Acs Catalysis, 10, 2020
|
|
6LO0
 
 | |
8D4Z
 
 | | Crystal structure of USP7 in complex with allosteric inhibitor FX1-3763 | | 分子名称: | 1-({(7M)-7-[1-(azetidin-3-yl)-6-chloro-1,2,3,4-tetrahydroquinolin-8-yl]thieno[3,2-b]pyridin-2-yl}methyl)pyrrolidine-2,5-dione, GLYCEROL, Ubiquitin carboxyl-terminal hydrolase 7 | | 著者 | Bell, J.A. | | 登録日 | 2022-06-03 | | 公開日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Novel USP7 inhibitors demonstrate potent anti-cancer activity in models of AML, synergy with BCL2 inhibition, and a differentiated mechanism of action
To Be Published
|
|
4X5Y
 
 | | Menin in complex with MI-503 | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-methyl-1-(1H-pyrazol-4-ylmethyl)-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, ... | | 著者 | Pollock, J, Borkin, D, Cierpicki, T, Grembecka, J. | | 登録日 | 2014-12-06 | | 公開日 | 2015-04-15 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Pharmacologic Inhibition of the Menin-MLL Interaction Blocks Progression of MLL Leukemia In Vivo.
Cancer Cell, 27, 2015
|
|
4X5Z
 
 | | menin in complex with MI-136 | | 分子名称: | 1,2-ETHANEDIOL, 5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, ... | | 著者 | Pollock, J, Borkin, D, Cierpicki, T, Grembecka, J. | | 登録日 | 2014-12-06 | | 公開日 | 2015-04-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Pharmacologic Inhibition of the Menin-MLL Interaction Blocks Progression of MLL Leukemia In Vivo.
Cancer Cell, 27, 2015
|
|
5UOT
 
 | |
5LJW
 
 | | MamK non-polymerising A278D mutant bound to AMPPNP | | 分子名称: | Actin-like ATPase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Lowe, J. | | 登録日 | 2016-07-20 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | X-ray and cryo-EM structures of monomeric and filamentous actin-like protein MamK reveal changes associated with polymerization.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
