7FEE
 
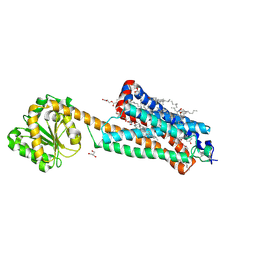 | | Crystal structure of the allosteric modulator ZCZ011 binding to CP55940-bound cannabinoid receptor 1 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, 6-methyl-3-[(1S)-2-nitro-1-thiophen-2-yl-ethyl]-2-phenyl-1H-indole, ... | | 著者 | Wang, X, Zhao, C, Shao, Z. | | 登録日 | 2021-07-19 | | 公開日 | 2022-06-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Molecular mechanism of allosteric modulation for the cannabinoid receptor CB1.
Nat.Chem.Biol., 18, 2022
|
|
4C7A
 
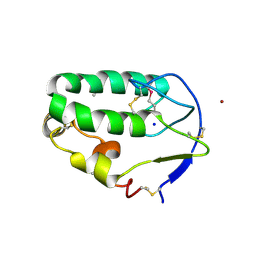 | | Crystal structure of the Smoothened CRD, selenomethionine-labeled | | 分子名称: | SMOOTHENED, SODIUM ION, ZINC ION | | 著者 | Nachtergaele, S, Whalen, D.M, Mydock, L.K, Zhao, Z, Malinauskas, T, Krishnan, K, Ingham, P.W, Covey, D.F, Rohatgi, R, Siebold, C. | | 登録日 | 2013-09-20 | | 公開日 | 2013-11-06 | | 最終更新日 | 2013-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure and Function of the Smoothened Extracellular Domain in Vertebrate Hedgehog Signaling
Elife, 2, 2013
|
|
4C79
 
 | | Crystal structure of the Smoothened CRD, native | | 分子名称: | SMOOTHENED, SODIUM ION, ZINC ION | | 著者 | Nachtergaele, S, Whalen, D.M, Mydock, L.K, Zhao, Z, Malinauskas, T, Krishnan, K, Ingham, P.W, Covey, D.F, Rohatgi, R, Siebold, C. | | 登録日 | 2013-09-20 | | 公開日 | 2013-11-06 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.604 Å) | | 主引用文献 | Structure and Function of the Smoothened Extracellular Domain in Vertebrate Hedgehog Signaling
Elife, 2, 2013
|
|
5J3J
 
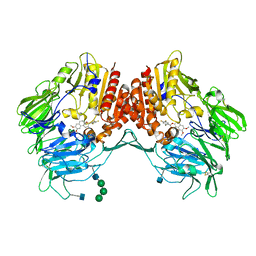 | | Crystal Structure of human DPP-IV in complex with HL1 | | 分子名称: | (2~{S},3~{R})-8,9-dimethoxy-3-[2,4,5-tris(fluoranyl)phenyl]-2,3-dihydro-1~{H}-benzo[f]chromen-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | 著者 | Wu, F, Li, H, Zhao, Z, Zhu, L, Xu, H, Li, S. | | 登録日 | 2016-03-31 | | 公開日 | 2017-04-05 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal Structure of human DPP-IV in complex with HL1
To Be Published
|
|
6JO7
 
 | | Crystal structure of mouse MXRA8 | | 分子名称: | Matrix remodeling-associated protein 8 | | 著者 | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, G.F. | | 登録日 | 2019-03-20 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
6JO8
 
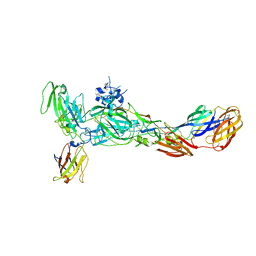 | | The complex structure of CHIKV envelope glycoprotein bound to human MXRA8 | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHIKV E1, ... | | 著者 | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, F.G. | | 登録日 | 2019-03-20 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.495 Å) | | 主引用文献 | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
4ZJC
 
 | | Structures of the human OX1 orexin receptor bound to selective and dual antagonists | | 分子名称: | OLEIC ACID, [5-(2-fluorophenyl)-2-methyl-1,3-thiazol-4-yl]{(2S)-2-[(5-phenyl-1,3,4-oxadiazol-2-yl)methyl]pyrrolidin-1-yl}methanone, human OX1R fusion protein to P.abysii glycogen synthase | | 著者 | Yin, J, Brautigam, C.A, Shao, Z, Clark, L, Harrell, C.M, Gotter, A.L, Coleman, P.J, Renger, J.J, Rosenbaum, D.M. | | 登録日 | 2015-04-29 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.832 Å) | | 主引用文献 | Structure and ligand-binding mechanism of the human OX1 and OX2 orexin receptors.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4ZJ8
 
 | | Structures of the human OX1 orexin receptor bound to selective and dual antagonists | | 分子名称: | OLEIC ACID, [(7R)-4-(5-chloro-1,3-benzoxazol-2-yl)-7-methyl-1,4-diazepan-1-yl][5-methyl-2-(2H-1,2,3-triazol-2-yl)phenyl]methanone, human OX1R fusion protein to P.abysii glycogen synthase | | 著者 | Yin, J, Brautigam, C.A, Shao, Z, Clark, L, Harrell, C.M, Gotter, A.L, Coleman, P, Renger, J.J, Rosenbaum, D.M. | | 登録日 | 2015-04-29 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.751 Å) | | 主引用文献 | Structure and ligand-binding mechanism of the human OX1 and OX2 orexin receptors.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6IH5
 
 | | Crystal structure of Phosphite Dehydrogenase mutant I151R/P176E from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine dinucleotide | | 分子名称: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Song, X, Feng, Y, Liu, Y, Zhao, Z. | | 登録日 | 2018-09-28 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.468 Å) | | 主引用文献 | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
6IH4
 
 | |
6IH2
 
 | |
6IH3
 
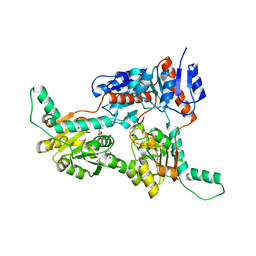 | | Crystal structure of Phosphite Dehydrogenase from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | 分子名称: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Song, X, Feng, Y, Zhao, Z, Liu, Y. | | 登録日 | 2018-09-28 | | 公開日 | 2019-03-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.942 Å) | | 主引用文献 | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
6IH8
 
 | |
6IH6
 
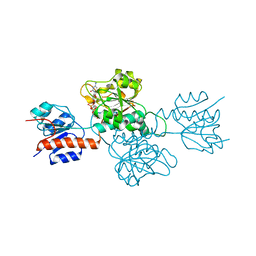 | | Phosphite Dehydrogenase mutant I151R/P176R/M207A from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine dinucleotide | | 分子名称: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Song, X, Feng, Y, Liu, Y, Zhao, Z. | | 登録日 | 2018-09-28 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.491 Å) | | 主引用文献 | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
7WV9
 
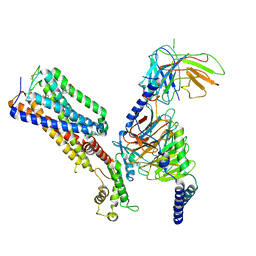 | | Allosteric modulator ZCZ011 binding to CP55940-bound cannabinoid receptor 1 in complex with Gi protein | | 分子名称: | 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, 6-methyl-3-[(1S)-2-nitro-1-thiophen-2-yl-ethyl]-2-phenyl-1H-indole, Cannabinoid receptor 1, ... | | 著者 | Xu, Z, Shao, Z. | | 登録日 | 2022-02-10 | | 公開日 | 2022-06-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.36 Å) | | 主引用文献 | Molecular mechanism of allosteric modulation for the cannabinoid receptor CB1.
Nat.Chem.Biol., 18, 2022
|
|
7F83
 
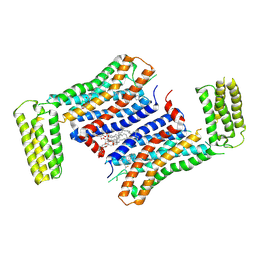 | | Crystal Structure of a receptor in Complex with inverse agonist | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(2-methylimidazo[2,1-b][1,3]thiazol-6-yl)-1-[2-[(1R)-5-(6-methylpyrimidin-4-yl)-2,3-dihydro-1H-inden-1-yl]-2,7-diazaspiro[3.5]nonan-7-yl]ethanone, Growth hormone secretagogue receptor type 1,Soluble cytochrome b562 | | 著者 | Xu, Z, Shao, Z. | | 登録日 | 2021-07-01 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Molecular mechanism of agonism and inverse agonism in ghrelin receptor.
Nat Commun, 13, 2022
|
|
7CKY
 
 | | Cryo-EM structure of PW0464 bound dopamine receptor DRD1-Gs signaling complex | | 分子名称: | 6-[4-[3-[bis(fluoranyl)methoxy]pyridin-2-yl]oxy-2-methyl-phenyl]-1,5-dimethyl-pyrimidine-2,4-dione, CHOLESTEROL, D(1A) dopamine receptor, ... | | 著者 | Yan, W, Shao, Z. | | 登録日 | 2020-07-20 | | 公開日 | 2021-03-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKZ
 
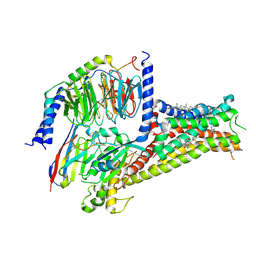 | | Cryo-EM structure of Dopamine and LY3154207 bound dopamine receptor DRD1-Gs signaling complex | | 分子名称: | 2-[2,6-bis(chloranyl)phenyl]-1-[(1S,3R)-3-(hydroxymethyl)-1-methyl-5-(3-methyl-3-oxidanyl-butyl)-3,4-dihydro-1H-isoquinolin-2-yl]ethanone, CHOLESTEROL, D(1A) dopamine receptor, ... | | 著者 | Yan, W, Shao, Z. | | 登録日 | 2020-07-20 | | 公開日 | 2021-03-03 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKX
 
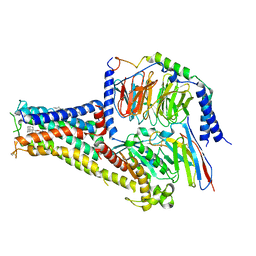 | | Cryo-EM structure of A77636 bound dopamine receptor DRD1-Gs signaling complex | | 分子名称: | (1R,3S)-3-(1-adamantyl)-1-(aminomethyl)-3,4-dihydro-1H-isochromene-5,6-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | 著者 | Yan, W, Shao, Z. | | 登録日 | 2020-07-20 | | 公開日 | 2021-03-03 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
8GQD
 
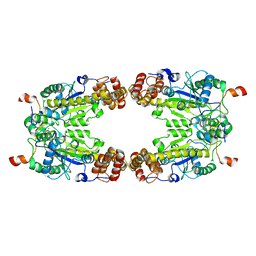 | | Complex Structure of Arginine Kinase McsB and McsA from Staphylococcus aureus | | 分子名称: | Protein-arginine kinase, Protein-arginine kinase activator protein, ZINC ION | | 著者 | Lu, K, Luo, B, Tao, X, Li, H, Xie, Y, Zhao, Z, Xia, W, Su, Z, Mao, Z. | | 登録日 | 2022-08-30 | | 公開日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.41 Å) | | 主引用文献 | Complex Structure and Activation Mechanism of Arginine Kinase McsB by McsA
To Be Published
|
|
6XAS
 
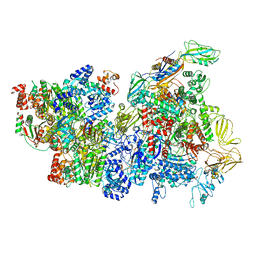 | | CryoEM Structure of E. coli Rho-dependent Transcription Pre-termination Complex | | 分子名称: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Hao, Z.T, Kim, H.K, Walz, T, Nudler, E. | | 登録日 | 2020-06-04 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Pre-termination Transcription Complex: Structure and Function.
Mol.Cell, 81, 2021
|
|
6XAV
 
 | | CryoEM Structure of E. coli Rho-dependent Transcription Pre-termination Complex bound with NusG | | 分子名称: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Hao, Z.T, Kim, H.K, Walz, T, Nudler, E. | | 登録日 | 2020-06-04 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (7.7 Å) | | 主引用文献 | Pre-termination Transcription Complex: Structure and Function.
Mol.Cell, 81, 2021
|
|
7UWH
 
 | | CryoEM Structure of E. coli Transcription-Coupled Ribonucleotide Excision Repair (TC-RER) complex bound to ribonucleotide substrate | | 分子名称: | DNA (59-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Hao, Z.T, Grower, M, Bharati, B, Proshkin, S, Epshtein, V, Svetlov, V, Nudler, E, Shamovsky, I. | | 登録日 | 2022-05-03 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | RNA polymerase drives ribonucleotide excision DNA repair in E. coli.
Cell, 186, 2023
|
|
7UWE
 
 | | CryoEM Structure of E. coli Transcription-Coupled Ribonucleotide Excision Repair (TC-RER) complex | | 分子名称: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Hao, Z.T, Grower, M, Bharati, B, Proshkin, S, Epshtein, V, Svetlov, V, Nudler, E, Shamovsky, I. | | 登録日 | 2022-05-03 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | RNA polymerase drives ribonucleotide excision DNA repair in E. coli.
Cell, 186, 2023
|
|
6JYV
 
 | | Structure of an isopenicillin N synthase from Pseudomonas aeruginosa PAO1 | | 分子名称: | Probable iron/ascorbate oxidoreductase, SODIUM ION | | 著者 | Hao, Z, Che, S, Wang, R, Liu, R, Zhang, Q, Bartlam, M. | | 登録日 | 2019-04-28 | | 公開日 | 2019-05-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.651 Å) | | 主引用文献 | Structural characterization of an isopenicillin N synthase family oxygenase from Pseudomonas aeruginosa PAO1.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
