6KVN
 
 | |
6KVO
 
 | | Crystal structure of chloroplast resolvase in complex with Holliday junction | | 分子名称: | DNA (5'-D(*AP*CP*AP*AP*CP*AP*GP*AP*TP*GP*AP*TP*GP*GP*AP*GP*CP*T)-3'), DNA (5'-D(*GP*CP*CP*TP*TP*GP*CP*TP*TP*GP*GP*AP*CP*AP*TP*CP*TP*T)-3'), DNA (5'-D(P*AP*AP*GP*AP*TP*GP*TP*CP*CP*AP*TP*CP*TP*GP*TP*TP*GP*T)-3'), ... | | 著者 | Yan, J.J, Hong, S.X, Guan, Z.Y, Yin, P. | | 登録日 | 2019-09-05 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1.
Nat Commun, 11, 2020
|
|
6LCT
 
 | | Crystal structure of catalytic inactive chloroplast resolvase NtMOC1 in complex with Holliday junction | | 分子名称: | DNA (5'-D(*AP*AP*GP*AP*TP*GP*TP*CP*CP*CP*TP*CP*TP*GP*TP*TP*GP*T)-3'), DNA (5'-D(*AP*CP*AP*AP*CP*AP*GP*AP*GP*GP*AP*TP*GP*GP*AP*GP*CP*T)-3'), DNA (5'-D(*GP*CP*CP*TP*TP*GP*CP*TP*GP*GP*GP*AP*CP*AP*TP*CP*TP*T)-3'), ... | | 著者 | Yan, J.J, Hong, S.X, Guan, Z.Y, Yin, P. | | 登録日 | 2019-11-19 | | 公開日 | 2020-04-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1.
Nat Commun, 11, 2020
|
|
8H0N
 
 | | Crystal structure of the human METTL1-WDR4 complex | | 分子名称: | tRNA (guanine-N(7)-)-methyltransferase, tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4 | | 著者 | Jin, X.H, Guan, Z.Y, Gong, Z, Zhang, D.L. | | 登録日 | 2022-09-30 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insight into how WDR4 promotes the tRNA N7-methylguanosine methyltransferase activity of METTL1.
Cell Discov, 9, 2023
|
|
5EKE
 
 | | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB (F215A mutant) | | 分子名称: | MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE, Uncharacterized glycosyltransferase sll0501 | | 著者 | Ardiccioni, C, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Liu, Q, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2015-11-03 | | 公開日 | 2016-01-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.001 Å) | | 主引用文献 | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB and insights into the mechanism of catalysis.
Nat Commun, 7, 2016
|
|
5EKP
 
 | | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB (WT) | | 分子名称: | MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE, Uncharacterized glycosyltransferase sll0501 | | 著者 | Ardiccioni, C, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Liu, Q, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2015-11-03 | | 公開日 | 2016-01-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.194 Å) | | 主引用文献 | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB and insights into the mechanism of catalysis.
Nat Commun, 7, 2016
|
|
3TPE
 
 | | The phipa p3121 structure | | 分子名称: | Serine/threonine-protein kinase HipA | | 著者 | Schumacher, M.A, Link, T, Brennan, R.G. | | 登録日 | 2011-09-07 | | 公開日 | 2012-10-03 | | 最終更新日 | 2018-03-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
7E44
 
 | | Crystal structure of NudC complexed with dpCoA | | 分子名称: | DEPHOSPHO COENZYME A, NADH pyrophosphatase, ZINC ION | | 著者 | Zhou, W, Guan, Z.Y, Yin, P, Zhang, D.L. | | 登録日 | 2021-02-10 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural insights into dpCoA-RNA decapping by NudC.
Rna Biol., 18, 2021
|
|
3TPB
 
 | | Structure of HipA(S150A) | | 分子名称: | CHLORIDE ION, PHOSPHATE ION, Serine/threonine-protein kinase HipA | | 著者 | schumacher, M.A. | | 登録日 | 2011-09-07 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
3TPD
 
 | | Structure of pHipA, monoclinic form | | 分子名称: | CHLORIDE ION, PHOSPHATE ION, Serine/threonine-protein kinase HipA | | 著者 | schumacher, M.A, link, T, Brennan, R.G. | | 登録日 | 2011-09-07 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
3TPT
 
 | | Structure of HipA(D309Q) bound to ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | 著者 | schumacher, M.A, link, T, Brennan, R.G. | | 登録日 | 2011-09-08 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
3TPV
 
 | |
8GQE
 
 | | Crystal structure of the W285A mutant of UVR8 in complex with RUP2 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ultraviolet-B receptor UVR8, WD repeat-containing protein RUP2 | | 著者 | Wang, Y.D, Wang, L.X, Guan, Z.Y, chang, H.F, Yin, P. | | 登録日 | 2022-08-30 | | 公開日 | 2022-09-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | RUP2 facilitates UVR8 redimerization via two interfaces.
Plant Commun., 4, 2023
|
|
6NC8
 
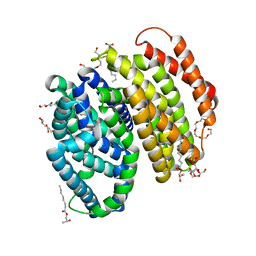 | | Lipid II flippase MurJ, inward occluded conformation | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Lipid II flippase MurJ, PENTAETHYLENE GLYCOL, ... | | 著者 | Kuk, A.C.Y, Lee, S.-Y. | | 登録日 | 2018-12-11 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.601 Å) | | 主引用文献 | Visualizing conformation transitions of the Lipid II flippase MurJ.
Nat Commun, 10, 2019
|
|
6NC9
 
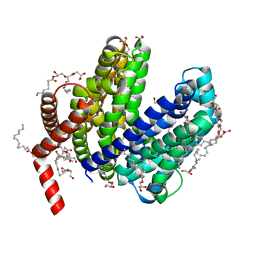 | | Lipid II flippase MurJ, outward-facing conformation | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Lipid II flippase MurJ, ... | | 著者 | Kuk, A.C.Y, Lee, S.-Y. | | 登録日 | 2018-12-11 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Visualizing conformation transitions of the Lipid II flippase MurJ.
Nat Commun, 10, 2019
|
|
6NC6
 
 | | Lipid II flippase MurJ, inward closed conformation | | 分子名称: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Lipid II flippase MurJ, ... | | 著者 | Kuk, A.C.Y, Lee, S.-Y. | | 登録日 | 2018-12-10 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Visualizing conformation transitions of the Lipid II flippase MurJ.
Nat Commun, 10, 2019
|
|
6NC7
 
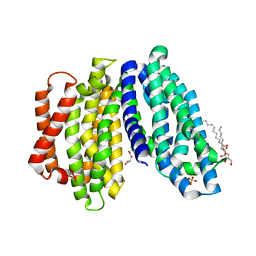 | |
5VXA
 
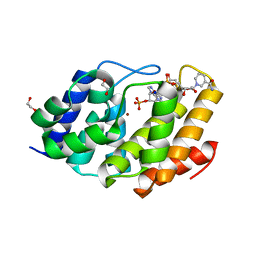 | | Structure of the human Mesh1-NADPH complex | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | 著者 | Rose, J, Zhou, P. | | 登録日 | 2017-05-23 | | 公開日 | 2018-05-23 | | 最終更新日 | 2022-03-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | MESH1 is a cytosolic NADPH phosphatase that regulates ferroptosis.
Nat Metab, 2, 2020
|
|
1JL3
 
 | | Crystal Structure of B. subtilis ArsC | | 分子名称: | ARSENATE REDUCTASE, SULFATE ION | | 著者 | Su, X.-D, Bennett, M.S. | | 登録日 | 2001-07-15 | | 公開日 | 2001-10-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Bacillus subtilis arsenate reductase is structurally and functionally similar to low molecular weight protein tyrosine phosphatases.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
8FHW
 
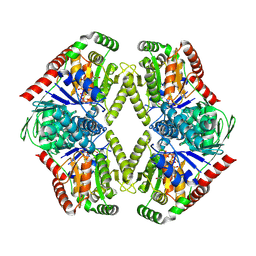 | |
8ITA
 
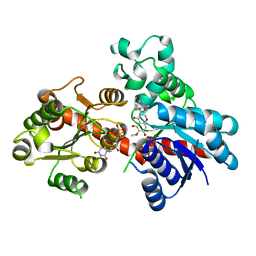 | | A reversible glycosyltransferase of tectorigenin - Bc7OUGT complexed with UDP and tectorigenin | | 分子名称: | 3-(4-hydroxyphenyl)-6-methoxy-5,7-bis(oxidanyl)chromen-4-one, Bc7OUGT, URIDINE-5'-DIPHOSPHATE | | 著者 | Zhang, Z.Y, Lu, L, Guan, Z.F, Cheng, W.J. | | 登録日 | 2023-03-22 | | 公開日 | 2023-08-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Functional characterization and structural basis of a reversible glycosyltransferase involves in plant chemical defence.
Plant Biotechnol J, 21, 2023
|
|
8Z9Z
 
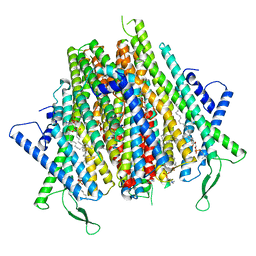 | | Cryo-EM structure of the insect olfactory receptor OR5-Orco heterocomplex from Acyrthosiphon pisum | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Odorant receptor, ApisOR5, ... | | 著者 | Wang, Y.D, Qiu, L, Guan, Z.Y, Wang, Q, Wang, G.R, Yin, P. | | 登録日 | 2024-04-24 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex.
Science, 384, 2024
|
|
8Z9A
 
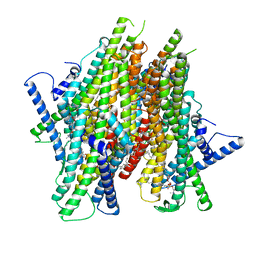 | | Cryo-EM structure of the insect olfactory receptor OR5-Orco heterocomplex from Acyrthosiphon pisum bound with geranyl acetate | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Odorant receptor, ApisOR5, ... | | 著者 | Wang, Y.D, Qiu, L, Guan, Z.Y, Wang, Q, Wang, G.R, Yin, P. | | 登録日 | 2024-04-23 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex.
Science, 384, 2024
|
|
8W5K
 
 | | Cryo-EM structure of the yeast TOM core complex crosslinked by BS3 (from TOM-TIM23 complex) | | 分子名称: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | 著者 | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | 登録日 | 2023-08-27 | | 公開日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
8W5J
 
 | | Cryo-EM structure of the yeast TOM core complex (from TOM-TIM23 complex) | | 分子名称: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | 著者 | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | 登録日 | 2023-08-27 | | 公開日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
