6KVF
 
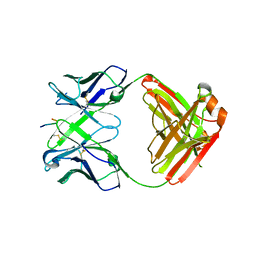 | | Structure of anti-hCXCR2 abN48 in complex with its CXCR2 epitope | | 分子名称: | Peptide from C-X-C chemokine receptor type 2, heavy chain, light chain | | 著者 | Xiang, J.C, Yan, L, Yang, B, Wilson, I.A. | | 登録日 | 2019-09-04 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Selection of a picomolar antibody that targets CXCR2-mediated neutrophil activation and alleviates EAE symptoms.
Nat Commun, 12, 2021
|
|
8JZS
 
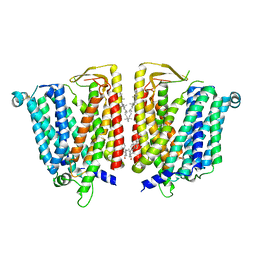 | | Outward-facing SLC15A4 dimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, lysosomal transporter | | 著者 | Zhang, S.S, Chen, X.D, Xie, M. | | 登録日 | 2023-07-06 | | 公開日 | 2023-09-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Structural basis for recruitment of TASL by SLC15A4 in human endolysosomal TLR signaling.
Nat Commun, 14, 2023
|
|
8JZU
 
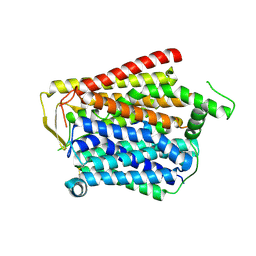 | | SLC15A4_TASL complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, TLR adapter,Green fluorescent protein, TSLAA-EGPF tag fusion protein | | 著者 | Zhang, S.S, Chen, X.D, Xie, M. | | 登録日 | 2023-07-06 | | 公開日 | 2023-09-27 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Structural basis for recruitment of TASL by SLC15A4 in human endolysosomal TLR signaling.
Nat Commun, 14, 2023
|
|
8JZR
 
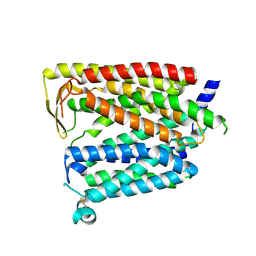 | | Outward_facing SLC15A4 monomer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, lysosomal transporter,ALFA tag | | 著者 | Zhang, S.S, Chen, X.D, Xie, M. | | 登録日 | 2023-07-06 | | 公開日 | 2023-09-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Structural basis for recruitment of TASL by SLC15A4 in human endolysosomal TLR signaling.
Nat Commun, 14, 2023
|
|
6AKW
 
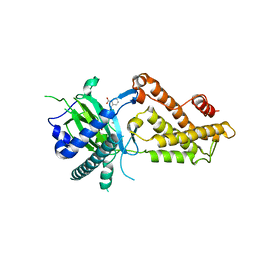 | | Crystal structure of RNA dioxygenase bound with an inhibitor | | 分子名称: | 2-OXOGLUTARIC ACID, 2-[[2,6-bis(chloranyl)-4-(3,5-dimethyl-1,2-oxazol-4-yl)phenyl]amino]benzoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO | | 著者 | Yang, C.-G, Huang, Y, Gan, J. | | 登録日 | 2018-09-04 | | 公開日 | 2019-05-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia.
Cancer Cell, 35, 2019
|
|
7UBF
 
 | |
7UBD
 
 | |
7UBI
 
 | |
7UCP
 
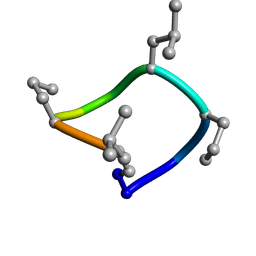 | | computationally designed macrocycle | | 分子名称: | computationally designed cyclic peptide D8.3.p2 | | 著者 | Bhardwaj, G, Baker, D, Rettie, S, Glynn, C, Sawaya, M. | | 登録日 | 2022-03-17 | | 公開日 | 2022-09-14 | | 最終更新日 | 2022-09-28 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Accurate de novo design of membrane-traversing macrocycles.
Cell, 185, 2022
|
|
7UZL
 
 | |
7UBC
 
 | |
7UBE
 
 | |
7UBG
 
 | |
7UBH
 
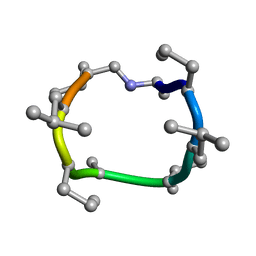 | |
3IKD
 
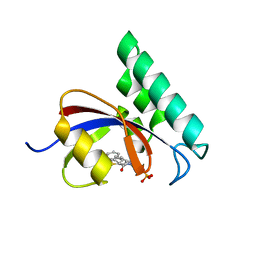 | | Structure-Based Design of Novel PIN1 Inhibitors (I) | | 分子名称: | (2R)-2-[(1-benzothiophen-2-ylcarbonyl)amino]-3-phenylpropyl phosphate, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Matthews, D, Greasley, S, Ferre, R, Parge, H. | | 登録日 | 2009-08-05 | | 公開日 | 2009-09-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-based design of novel human Pin1 inhibitors (I).
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3IK8
 
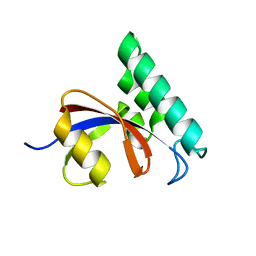 | |
3IKG
 
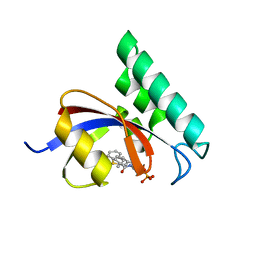 | | Structure-Based Design of Novel PIN1 Inhibitors (I) | | 分子名称: | (2R)-2-[(1-benzothiophen-2-ylcarbonyl)amino]-3-(3-methylphenyl)propyl phosphate, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Parge, H, Ferre, R.A, Greasley, S, Matthews, D. | | 登録日 | 2009-08-05 | | 公開日 | 2009-09-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Structure-based design of novel human Pin1 inhibitors (I).
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5W1R
 
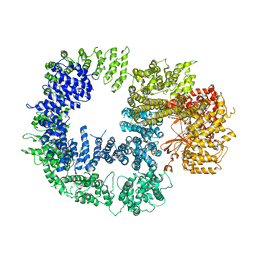 | | Cryo-EM structure of DNAPKcs | | 分子名称: | DNA-dependent protein kinase catalytic subunit | | 著者 | Sharif, H, Li, Y, Wu, H. | | 登録日 | 2017-06-04 | | 公開日 | 2017-07-19 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Cryo-EM structure of the DNA-PK holoenzyme.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8OOR
 
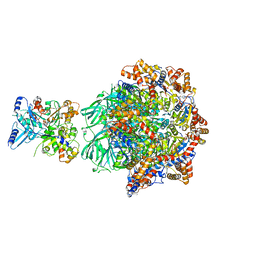 | | CryoEM Structure INO80core Hexasome complex Rvb core refinement state2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.87 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOT
 
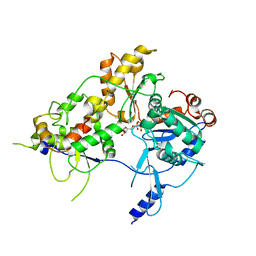 | | CryoEM Structure INO80core Hexasome complex Arp5 Ies6 refinement state2 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling complex subunit IES6, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOS
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-hexasome refinement state 2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase Ino80, DNA Strand 2, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
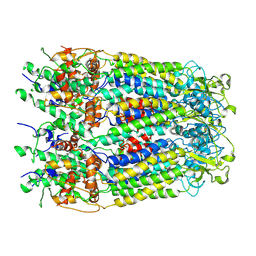
8OOC
 
 | | CryoEM Structure INO80core Hexasome complex Rvb core refinement state1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chromatin-remodeling ATPase Ino80, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
5VFI
 
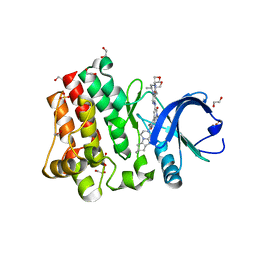 | | Bruton's tyrosine kinase (BTK) with GDC-0853 | | 分子名称: | 1,2-ETHANEDIOL, 2-[3'-(hydroxymethyl)-1-methyl-5-({5-[(2S)-2-methyl-4-(oxetan-3-yl)piperazin-1-yl]pyridin-2-yl}amino)-6-oxo[1,6-dihydro[3,4'-bipyridine]]-2'-yl]-7,7-dimethyl-3,4,7,8-tetrahydro-2H-cyclopenta[4,5]pyrrolo[1,2-a]pyrazin-1(6H)-one, SULFATE ION, ... | | 著者 | Steinbacher, S, Eigenbrot, C. | | 登録日 | 2017-04-07 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Discovery of GDC-0853: A Potent, Selective, and Noncovalent Bruton's Tyrosine Kinase Inhibitor in Early Clinical Development.
J. Med. Chem., 61, 2018
|
|
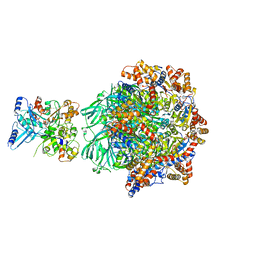
6M02
 
 | | cryo-EM structure of human Pannexin 1 channel | | 分子名称: | Pannexin-1 | | 著者 | Ronggui, Q, Lili, D, Jilin, Z, Xuekui, Y, Lei, W, Shujia, Z. | | 登録日 | 2020-02-19 | | 公開日 | 2020-03-25 | | 最終更新日 | 2020-05-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structure of human heptameric Pannexin 1 channel.
Cell Res., 30, 2020
|
|
4GEH
 
 | |
