6OI1
 
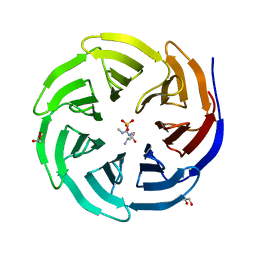 | | Crystal structure of human WDR5 in complex with monomethyl L-arginine | | 分子名称: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, GLYCEROL, SULFATE ION, ... | | 著者 | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | 登録日 | 2019-04-08 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
4X4X
 
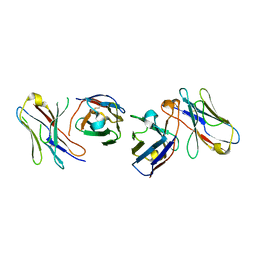 | |
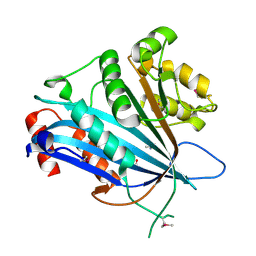
7BXW
 
 | |
4X6H
 
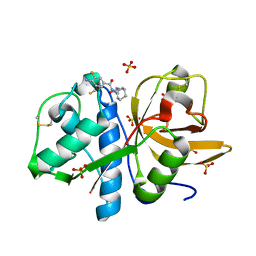 | | Development of N-(Functionalized benzoyl)-homocycloleucyl-glycinonitriles as Potent Cathepsin K Inhibitors. | | 分子名称: | 4-amino-3-fluoro-N-(1-{[(2Z)-2-iminoethyl]carbamoyl}cyclohexyl)benzamide, 4-amino-N-{1-[(cyanomethyl)carbamoyl]cyclohexyl}-3-fluorobenzamide, Cathepsin K, ... | | 著者 | Borisek, J, Mohar, B, Vizovisek, M, Sosnowski, P, Turk, D, Turk, B, Novic, M. | | 登録日 | 2014-12-08 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Development of N-(Functionalized benzoyl)-homocycloleucyl-glycinonitriles as Potent Cathepsin K Inhibitors.
J.Med.Chem., 58, 2015
|
|
1GP9
 
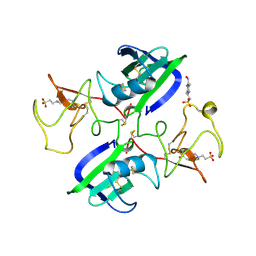 | | A New Crystal Form of the Nk1 Splice Variant of Hgf/Sf Demonstrates Extensive Hinge Movement and Suggests that the Nk1 Dimer Originates by Domain Swapping | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEPATOCYTE GROWTH FACTOR | | 著者 | Watanabe, K, Chirgadze, D.Y, Lietha, D, Gherardi, E, Blundell, T.L. | | 登録日 | 2001-10-31 | | 公開日 | 2001-11-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A New Crystal Form of the Nk1 Splice Variant of Hgf/Sf Demonstrates Extensive Hinge Movement and Suggests that the Nk1 Dimer Originates by Domain Swapping
J.Mol.Biol., 319, 2002
|
|
4WFB
 
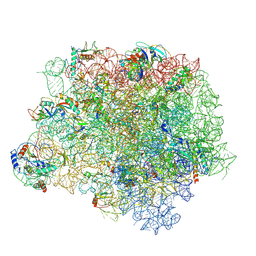 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with BC-3205 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S rRNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Eyal, Z, Matzov, D, Krupkin, M, Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A.E. | | 登録日 | 2014-09-14 | | 公開日 | 2015-10-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.43 Å) | | 主引用文献 | Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6GCH
 
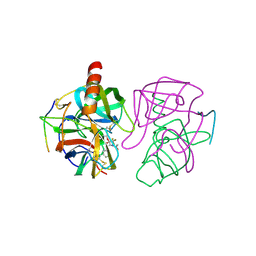 | | STRUCTURE OF CHYMOTRYPSIN-*TRIFLUOROMETHYL KETONE INHIBITOR COMPLEXES. COMPARISON OF SLOWLY AND RAPIDLY EQUILIBRATING INHIBITORS | | 分子名称: | 1,1,1-TRIFLUORO-3-ACETAMIDO-4-PHENYL BUTAN-2-ONE(N-ACETYL-L-PHENYLALANYL TRIFLUOROMETHYL KETONE), GAMMA-CHYMOTRYPSIN A | | 著者 | Brady, K, Wei, A, Ringe, D, Abeles, R.H. | | 登録日 | 1990-04-06 | | 公開日 | 1990-10-15 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of chymotrypsin-trifluoromethyl ketone inhibitor complexes: comparison of slowly and rapidly equilibrating inhibitors.
Biochemistry, 29, 1990
|
|
4WMF
 
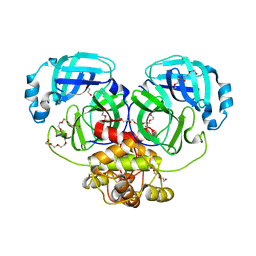 | | Crystal structure of catalytically inactive MERS-CoV 3CL protease (C148A) in spacegroup P212121 | | 分子名称: | DI(HYDROXYETHYL)ETHER, MERS-CoV 3CL protease, TETRAETHYLENE GLYCOL | | 著者 | Lountos, G.T, Needle, D, Waugh, D.S. | | 登録日 | 2014-10-08 | | 公開日 | 2015-05-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structures of the Middle East respiratory syndrome coronavirus 3C-like protease reveal insights into substrate specificity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4X09
 
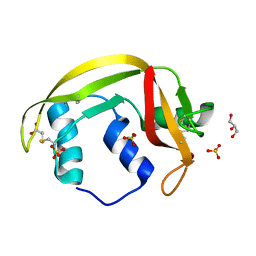 | | Structure of human RNase 6 in complex with sulphate anions | | 分子名称: | GLYCEROL, Ribonuclease K6, SULFATE ION | | 著者 | Prats-Ejarque, G, Arranz-Trullen, J, Blanco, J.A, Pulido, D, Moussaoui, M, Boix, E. | | 登録日 | 2014-11-21 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.722 Å) | | 主引用文献 | The first crystal structure of human RNase 6 reveals a novel substrate-binding and cleavage site arrangement.
Biochem.J., 473, 2016
|
|
6GF0
 
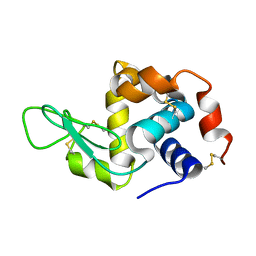 | | Lysozyme structure determined from SFX data using a Sheet-on-Sheet chipless chip | | 分子名称: | Lysozyme C | | 著者 | Doak, R.B, Gorel, A, Foucar, L, Gruenbein, M.L, Hilpert, M, Kloos, M, Nass Kovacs, G, Roome, C, Shoeman, R.L, Stricker, M, Tono, K, You, D, Ueda, K, Sherrel, D, Owen, R, Barends, T.R.M, Schlichting, I. | | 登録日 | 2018-04-27 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Crystallography on a chip - without the chip: sheet-on-sheet sandwich.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
8FUU
 
 | |
6PVK
 
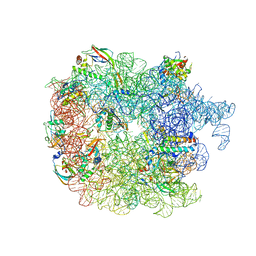 | | Bacterial 45SRbgA ribosomal particle class A | | 分子名称: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | 著者 | Ortega, J, Seffouh, A, Jain, N, Jahagirdar, D, Basu, K, Razi, A, Ni, X, Guarne, A, Britton, R.A. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural consequences of the interaction of RbgA with a 50S ribosomal subunit assembly intermediate.
Nucleic Acids Res., 47, 2019
|
|
7B4M
 
 | |
4X67
 
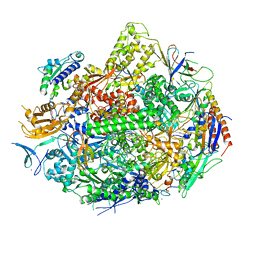 | | Crystal structure of elongating yeast RNA polymerase II stalled at oxidative Cyclopurine DNA lesions. | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | 著者 | Wang, L, Chong, J, Wang, D. | | 登録日 | 2014-12-07 | | 公開日 | 2015-02-04 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (4.1 Å) | | 主引用文献 | Mechanism of RNA polymerase II bypass of oxidative cyclopurine DNA lesions.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7B4L
 
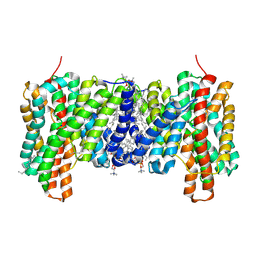 | |
4X6Z
 
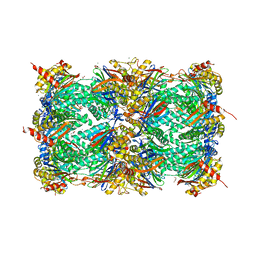 | | Yeast 20S proteasome in complex with PR-VI modulator | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Rostankowski, R, Witkowska, J, Borek, D, Otwinowski, Z, Jankowska, E. | | 登録日 | 2014-12-09 | | 公開日 | 2015-12-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structures revealed the common place of binding of low-molecular
mass activators with the 20S proteasome
To Be Published
|
|
3SGP
 
 | | Amyloid-related segment of alphaB-crystallin residues 90-100 mutant V91L | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha-crystallin B chain | | 著者 | Laganowsky, A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | 登録日 | 2011-06-15 | | 公開日 | 2012-03-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.4016 Å) | | 主引用文献 | Atomic view of a toxic amyloid small oligomer.
Science, 335, 2012
|
|
1GXC
 
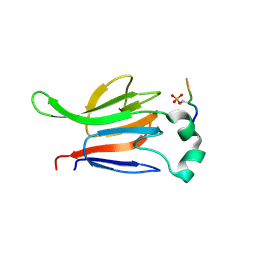 | | FHA domain from human Chk2 kinase in complex with a synthetic phosphopeptide | | 分子名称: | SERINE/THREONINE-PROTEIN KINASE CHK2, SYNTHETIC PHOSPHOPEPTIDE | | 著者 | Li, J, Williams, B.L, Haire, L.F, Goldberg, M, Wilker, E, Durocher, D, Yaffe, M.B, Jackson, S.P, Smerdon, S.J. | | 登録日 | 2002-04-02 | | 公開日 | 2002-06-13 | | 最終更新日 | 2016-12-21 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural and Functional Versatility of the Fha Domain in DNA-Damage Signaling by the Tumor Suppressor Kinase Chk2
Mol.Cell, 9, 2002
|
|
6Q30
 
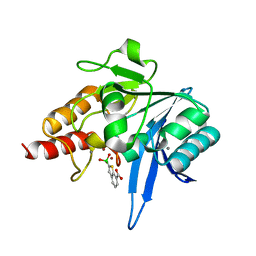 | | Crystal structure of NDM-1 beta-lactamase in complex with boronic inhibitor cpd 5 | | 分子名称: | (7-carboxy-1-benzothiophen-2-yl)-tris(oxidanyl)boranuide, CALCIUM ION, Metallo-beta-lactamase type 2, ... | | 著者 | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Celenza, G, Venturelli, A, Costi, M.P, Tondi, D, Cendron, L. | | 登録日 | 2018-12-03 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6Q3E
 
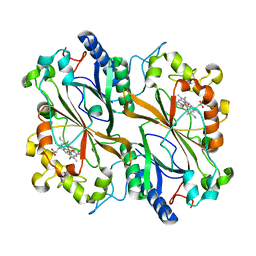 | | Dye type peroxidase Aa from Streptomyces lividans: 274.4 kGy structure | | 分子名称: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Ebrahim, A, Moreno-Chicano, T, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Appleby, M, Owen, R.L. | | 登録日 | 2018-12-04 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins.
Iucrj, 6, 2019
|
|
1GO5
 
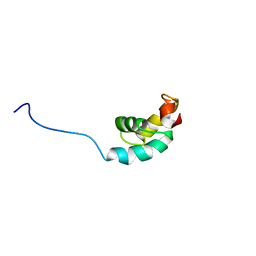 | |
4X6A
 
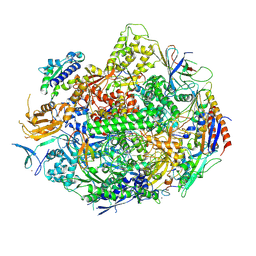 | | Crystal structure of yeast RNA polymerase II encountering oxidative Cyclopurine DNA lesions | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | 著者 | Wang, L, Chong, J, Wang, D. | | 登録日 | 2014-12-07 | | 公開日 | 2015-02-04 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (3.96 Å) | | 主引用文献 | Mechanism of RNA polymerase II bypass of oxidative cyclopurine DNA lesions.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1UXD
 
 | | Fructose repressor DNA-binding domain, NMR, 34 structures | | 分子名称: | FRUCTOSE REPRESSOR | | 著者 | Penin, F, Geourjon, C, Montserret, R, Bockmann, A, Lesage, A, Yang, Y, Bonod-Bidaud, C, Cortay, J.C, Negre, D, Cozzone, A.J, Deleage, G. | | 登録日 | 1996-12-26 | | 公開日 | 1997-04-01 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional structure of the DNA-binding domain of the fructose repressor from Escherichia coli by 1H and 15N NMR.
J.Mol.Biol., 270, 1997
|
|
6Q6G
 
 | | Cryo-EM structure of the APC/C-Cdc20-Cdk2-cyclinA2-Cks2 complex, the D1 box class | | 分子名称: | Anaphase-promoting complex subunit 1,Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | 著者 | Zhang, S, Barford, D. | | 登録日 | 2018-12-11 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cyclin A2 degradation during the spindle assembly checkpoint requires multiple binding modes to the APC/C.
Nat Commun, 10, 2019
|
|
4X6I
 
 | | Development of N-(Functionalized benzoyl)-homocycloleucyl-glycinonitriles as Potent Cathepsin K Inhibitors. | | 分子名称: | 2-amino-4-bromo-N-{1-[(cyanomethyl)carbamoyl]cyclohexyl}benzamide, Cathepsin K, SULFATE ION | | 著者 | Borisek, J, Mohar, B, Vizovisek, M, Sosnowski, P, Turk, D, Turk, B, Novic, M. | | 登録日 | 2014-12-08 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Development of N-(Functionalized benzoyl)-homocycloleucyl-glycinonitriles as Potent Cathepsin K Inhibitors.
J.Med.Chem., 58, 2015
|
|
