7L58
 
 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab H4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab H4 variable domain heavy chain, ... | | 著者 | Rapp, M, Shapiro, L. | | 登録日 | 2020-12-21 | | 公開日 | 2021-04-14 | | 最終更新日 | 2021-04-21 | | 実験手法 | ELECTRON MICROSCOPY (5.07 Å) | | 主引用文献 | Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class.
Cell Rep, 35, 2021
|
|
7CPO
 
 | | Crystal Structure of Anolis carolinensis MHC I complex | | 分子名称: | HIS-VAL-TYR-GLY-PRO-LEU-LYS-PRO-ILE, MHC CLASS I ANTIGEN, beta2-microglobulin | | 著者 | Wang, Y, Qu, Z, Ma, L, Wei, X, Zhang, N, Xia, C. | | 登録日 | 2020-08-07 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Crystal Structure of the MHC Class I (MHC-I) Molecule in the Green Anole Lizard Demonstrates the Unique MHC-I System in Reptiles.
J Immunol., 206, 2021
|
|
7LLK
 
 | |
7KMB
 
 | | ACE2-RBD Focused Refinement Using Symmetry Expansion of Applied C3 for Triple ACE2-bound SARS-CoV-2 Trimer Spike at pH 7.4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-02 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KNH
 
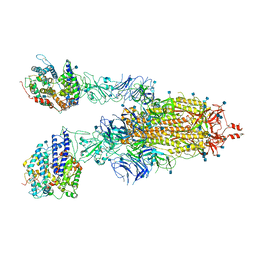 | | Cryo-EM Structure of Double ACE2-Bound SARS-CoV-2 Trimer Spike at pH 5.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-04 | | 公開日 | 2020-12-16 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.74 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7LY9
 
 | |
7LZE
 
 | | Cryo-EM Structure of disulfide stabilized HMPV F v4-B | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0 | | 著者 | Gorman, J, Kwong, P.D. | | 登録日 | 2021-03-09 | | 公開日 | 2021-08-25 | | 最終更新日 | 2021-10-06 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | Interprotomer disulfide-stabilized variants of the human metapneumovirus fusion glycoprotein induce high titer-neutralizing responses.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LG6
 
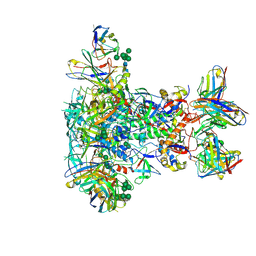 | | BG505 SOSIP.v5.2 in complex with VRC40.01 and RM19R Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | 著者 | Cottrell, C.A, Torres, J.L, Wu, N.R, Ward, A.B. | | 登録日 | 2021-01-19 | | 公開日 | 2021-09-15 | | 最終更新日 | 2021-11-17 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Structural basis of glycan276-dependent recognition by HIV-1 broadly neutralizing antibodies.
Cell Rep, 37, 2021
|
|
7LPN
 
 | |
5LY0
 
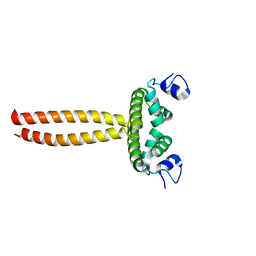 | | Crystal structure of LOB domain of Ramosa2 from Wheat | | 分子名称: | LOB family transfactor Ramosa2.1, ZINC ION | | 著者 | Wei, X.-B, Zhang, B, Chen, W.-F, Fan, S.-H, Rety, S, Xi, X.-G. | | 登録日 | 2016-09-23 | | 公開日 | 2017-10-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.877 Å) | | 主引用文献 | Structural analysis reveals a "molecular calipers" mechanism for a LATERAL ORGAN BOUNDARIES DOMAIN transcription factor protein from wheat.
J. Biol. Chem., 294, 2019
|
|
7E5X
 
 | |
4M58
 
 | | Crystal Structure of an transition metal transporter | | 分子名称: | Cobalamin biosynthesis protein CbiM, NICKEL (II) ION | | 著者 | Yu, Y, Yan, C.Y, Zhang, B, Li, X.L, Gu, J.K. | | 登録日 | 2013-08-08 | | 公開日 | 2014-03-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.21 Å) | | 主引用文献 | Planar substrate-binding site dictates the specificity of ECF-type nickel/cobalt transporters
Cell Res., 24, 2014
|
|
7DVP
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp4|5 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp4/5 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-14 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW0
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp14|15 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp14/15 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW6
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp15|16 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp15/16 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVY
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp9|10 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp9/10 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVX
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp6|7 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp6/7 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVW
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp5|6 peptidyl substrate | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, nsp5/6 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4JPW
 
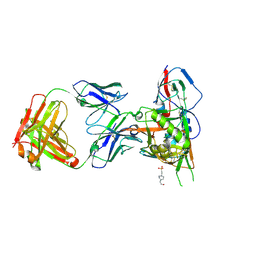 | | Crystal structure of broadly and potently neutralizing antibody 12a21 in complex with hiv-1 strain 93th057 gp120 mutant | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEAVY CHAIN OF ANTIBODY 12A21, ... | | 著者 | Acharya, P, Luongo, T, Zhou, T, Kwong, P.D. | | 登録日 | 2013-03-19 | | 公開日 | 2013-04-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.904 Å) | | 主引用文献 | Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization.
Cell(Cambridge,Mass.), 153, 2013
|
|
7WU3
 
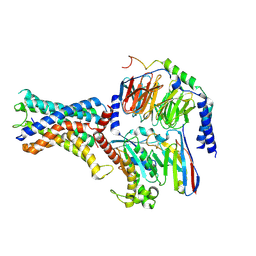 | | Cryo-EM structure of the adhesion GPCR ADGRF1 in complex with miniGs | | 分子名称: | Adhesion G-protein coupled receptor F1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | 登録日 | 2022-02-05 | | 公開日 | 2022-04-27 | | 最終更新日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7WU2
 
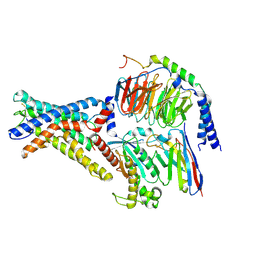 | | Cryo-EM structure of the adhesion GPCR ADGRD1 in complex with miniGs | | 分子名称: | Adhesion G-protein coupled receptor D1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | 登録日 | 2022-02-05 | | 公開日 | 2022-04-27 | | 最終更新日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7WU4
 
 | | Cryo-EM structure of the adhesion GPCR ADGRF1 in complex with miniGi | | 分子名称: | Adhesion G-protein coupled receptor F1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | 登録日 | 2022-02-05 | | 公開日 | 2022-04-27 | | 最終更新日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7WU5
 
 | | Cryo-EM structure of the adhesion GPCR ADGRF1(H565A/T567A) in complex with miniGi | | 分子名称: | Adhesion G-protein coupled receptor F1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | 登録日 | 2022-02-05 | | 公開日 | 2022-04-27 | | 最終更新日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
6KUU
 
 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3) | | 分子名称: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | 著者 | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | 登録日 | 2019-09-02 | | 公開日 | 2019-12-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3)
To Be Published
|
|
7D6M
 
 | |
