7ZO6
 
 | | L1 metallo-beta-lactamase in complex with hydrolysed cefoxitin | | 分子名称: | (2~{R},5~{S})-5-(aminocarbonyloxymethyl)-2-[(1~{S})-1-methoxy-2-oxidanyl-2-oxidanylidene-1-(2-thiophen-2-ylethanoylamino)ethyl]-5,6-dihydro-2~{H}-1,3-thiazine-4-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | 著者 | Hinchliffe, P, Spencer, J. | | 登録日 | 2022-04-24 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Interactions of hydrolyzed beta-lactams with the L1 metallo-beta-lactamase: Crystallography supports stereoselective binding of cephem/carbapenem products.
J.Biol.Chem., 299, 2023
|
|
7ZO7
 
 | | L1 metallo-beta-lactamase in complex with hydrolysed cefmetazole | | 分子名称: | (2R,5R)-2-[(1S)-1-[2-(cyanomethylsulfanyl)ethanoylamino]-1-methoxy-2-oxidanyl-2-oxidanylidene-ethyl]-5-[(1-methyl-1,2,3,4-tetrazol-5-yl)sulfanylmethyl]-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | 著者 | Hinchliffe, P, Spencer, J. | | 登録日 | 2022-04-24 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Interactions of hydrolyzed beta-lactams with the L1 metallo-beta-lactamase: Crystallography supports stereoselective binding of cephem/carbapenem products.
J.Biol.Chem., 299, 2023
|
|
5LRN
 
 | |
5NDB
 
 | | Crystal structure of metallo-beta-lactamase SPM-1 complexed with cyclobutanone inhibitor | | 分子名称: | (1~{S},4~{R},5~{S})-7,7-bis(chloranyl)-6,6-bis(oxidanyl)-2$l^{4}-thiabicyclo[3.2.0]hept-2-ene-4-carboxylic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase IMP-1, ... | | 著者 | Hinchliffe, P, Spencer, J. | | 登録日 | 2017-03-08 | | 公開日 | 2018-01-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Cyclobutanone Mimics of Intermediates in Metallo-beta-Lactamase Catalysis.
Chemistry, 24, 2018
|
|
5NE3
 
 | |
5NDE
 
 | |
4EZH
 
 | |
4EZ4
 
 | | free KDM6B structure | | 分子名称: | Lysine-specific demethylase 6B, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | 著者 | Cheng, Z.J, Patel, D.J. | | 登録日 | 2012-05-02 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response.
Nature, 488, 2012
|
|
4EYU
 
 | |
2XLN
 
 | | Crystal structure of a complex between Actinomadura R39 DD-peptidase and a boronate inhibitor | | 分子名称: | COBALT (II) ION, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE,, SULFATE ION, ... | | 著者 | Sauvage, E, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | 登録日 | 2010-07-21 | | 公開日 | 2011-01-26 | | 最終更新日 | 2014-06-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure Guided Development of Potent Reversibly Binding Penicillin Binding Protein Inhibitors
Acs Med.Chem.Lett., 2, 2011
|
|
2WPW
 
 | | Tandem GNAT protein from the clavulanic acid biosynthesis pathway (without AcCoA) | | 分子名称: | ACETYL COENZYME *A, ORF14 | | 著者 | Iqbal, A, Arunlanantham, H, McDonough, M.A, Chowdhury, R, Clifton, I.J. | | 登録日 | 2009-08-11 | | 公開日 | 2009-12-29 | | 最終更新日 | 2018-03-28 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Crystallographic and mass spectrometric analyses of a tandem GNAT protein from the clavulanic acid biosynthesis pathway.
Proteins, 78, 2010
|
|
2XK1
 
 | | Crystal structure of a complex between Actinomadura R39 DD-peptidase and a boronate inhibitor | | 分子名称: | COBALT (II) ION, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, SULFATE ION, ... | | 著者 | Sauvage, E, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | 登録日 | 2010-07-07 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure Guided Development of Potent Reversibly Binding Penicillin Binding Protein Inhibitors
Acs Med.Chem.Lett., 2, 2011
|
|
2XML
 
 | | Crystal structure of human JMJD2C catalytic domain | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, LYSINE-SPECIFIC DEMETHYLASE 4C, ... | | 著者 | Yue, W.W, Gileadi, C, Krojer, T, Pike, A.C.W, von Delft, F, Ng, S, Carpenter, L, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | 登録日 | 2010-07-28 | | 公開日 | 2010-09-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural and Evolutionary Basis for the Dual Substrate Selectivity of Human Kdm4 Histone Demethylase Family.
J.Biol.Chem., 286, 2011
|
|
2XUE
 
 | | CRYSTAL STRUCTURE OF JMJD3 | | 分子名称: | 2-OXOGLUTARIC ACID, FE (III) ION, LYSINE-SPECIFIC DEMETHYLASE 6B, ... | | 著者 | Chung, C, Rowland, P, Mosley, J, Thomas, P.J. | | 登録日 | 2010-10-19 | | 公開日 | 2011-12-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A Selective Jumonji H3K27 Demethylase Inhibitor Modulates the Proinflammatory Macrophage Response
Nature, 488, 2012
|
|
2XDV
 
 | | Crystal Structure of the Catalytic Domain of FLJ14393 | | 分子名称: | 1,2-ETHANEDIOL, CADMIUM ION, MANGANESE (II) ION, ... | | 著者 | Krojer, T, Muniz, J.R.C, Ng, S.S, Pilka, E, Guo, K, Pike, A.C.W, Filippakopoulos, P, Knapp, S, Kavanagh, K.L, Gileadi, O, Bunkoczi, G, Yue, W.W, Niesen, F, Sobott, F, Fedorov, O, Savitsky, P, Kochan, G, Daniel, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U. | | 登録日 | 2010-05-07 | | 公開日 | 2010-05-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
2WPX
 
 | | Tandem GNAT protein from the clavulanic acid biosynthesis pathway (with AcCoA) | | 分子名称: | ACETYL COENZYME *A, GLYCEROL, ORF14 | | 著者 | Iqbal, A, Arunlanantham, H, McDonough, M.A, Chowdhury, R, Clifton, I.J. | | 登録日 | 2009-08-11 | | 公開日 | 2009-12-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Crystallographic and mass spectrometric analyses of a tandem GNAT protein from the clavulanic acid biosynthesis pathway.
Proteins, 78, 2010
|
|
2X71
 
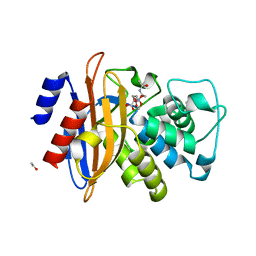 | | Structural basis for the interaction of lactivicins with serine beta- lactamases | | 分子名称: | (2E)-2-{[(2S)-2-(ACETYLAMINO)-2-CARBOXYETHOXY]IMINO}PENTANEDIOIC ACID, BETA-LACTAMASE, ETHANOL, ... | | 著者 | Sauvage, E, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | 登録日 | 2010-02-22 | | 公開日 | 2010-07-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis for the Interaction of Lactivicins with Serine Beta-Lactamases.
J.Med.Chem., 53, 2010
|
|
1QNJ
 
 | | THE STRUCTURE OF NATIVE PORCINE PANCREATIC ELASTASE AT ATOMIC RESOLUTION (1.1 A) | | 分子名称: | ELASTASE, SODIUM ION, SULFATE ION | | 著者 | Wurtele, M, Hahn, M, Hilpert, K, Hohne, W. | | 登録日 | 1999-10-15 | | 公開日 | 2000-03-31 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Atomic Resolution Structure of Native Porcine Pancreatic Elastase at 1.1 A
Acta Crystallogr.,Sect.D, 56, 2000
|
|
7GJX
 
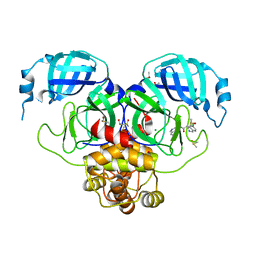 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-3ccb8ef6-1 (Mpro-P0744) | | 分子名称: | (4S)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GKE
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-dd3ad2b5-3 (Mpro-P0851) | | 分子名称: | (4S)-6-chloro-N~4~-(isoquinolin-4-yl)-3,4-dihydroisoquinoline-2,4(1H)-dicarboxamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GKU
 
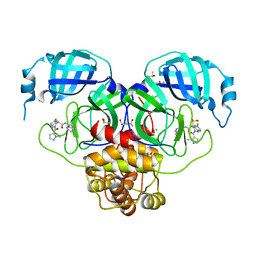 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-4223bc15-40 (Mpro-P1079) | | 分子名称: | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-(1-methyl-1H-pyrazole-5-carbonyl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.866 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GLA
 
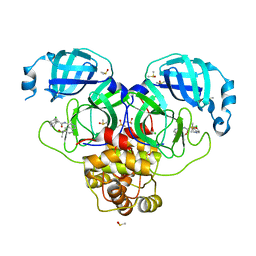 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-86c60949-2 (Mpro-P1812) | | 分子名称: | (4R)-6-chloro-N-[6-(2-hydroxypropan-2-yl)isoquinolin-4-yl]-1,2,3,4-tetrahydroquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.684 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GLQ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with PET-UNK-8df914d1-2 (Mpro-P2007) | | 分子名称: | 2-(3-chlorophenyl)-N-[(4R)-imidazo[1,2-a]pyridin-3-yl]acetamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.063 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GM5
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-5cd9ea36-2 (Mpro-P2089) | | 分子名称: | (4S)-6-chloro-2-(cyclopropylsulfamoyl)-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.709 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GMK
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with PET-UNK-d899bab6-1 (Mpro-P2201) | | 分子名称: | 2-(3-chlorophenyl)-N-[6-(dimethylamino)isoquinolin-4-yl]acetamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.845 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
