2C3O
 
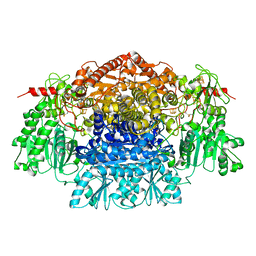 | | CRYSTAL STRUCTURE OF THE FREE RADICAL INTERMEDIATE OF PYRUVATE:FERREDOXIN OXIDOREDUCTASE FROM Desulfovibrio africanus | | 分子名称: | CALCIUM ION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | 著者 | Cavazza, C, Contreras-Martel, C, Pieulle, L, Chabriere, E, Hatchikian, E.C, Fontecilla-Camps, J.C. | | 登録日 | 2005-10-11 | | 公開日 | 2006-02-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Flexibility of Thiamine Diphosphate Revealed by Kinetic Crystallographic Studies of the Reaction of Pyruvate-Ferredoxin Oxidoreductase with Pyruvate.
Structure, 14, 2006
|
|
2C3U
 
 | | Crystal Structure Of Pyruvate-Ferredoxin Oxidoreductase From Desulfovibrio africanus, Oxygen inhibited form | | 分子名称: | 2-(3-{[4-(HYDROXYAMINO)-2-METHYLPYRIMIDIN-5-YL]METHYL}-4-METHYL-2,3-DIHYDRO-1,3-THIAZOL-5-YL)ETHYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, IRON/SULFUR CLUSTER, ... | | 著者 | Cavazza, C, Contreras-Martel, C, Pieulle, L, Chabriere, E, Hatchikian, E.C, Fontecilla-Camps, J.C. | | 登録日 | 2005-10-12 | | 公開日 | 2006-02-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Flexibility of Thiamine Diphosphate Revealed by Kinetic Crystallographic Studies of the Reaction of Pyruvate-Ferredoxin Oxidoreductase with Pyruvate.
Structure, 14, 2006
|
|
1R4G
 
 | | Solution structure of the Sendai virus protein X C-subdomain | | 分子名称: | RNA polymerase alpha subunit | | 著者 | Blanchard, L, Tarbouriech, N, Blackledge, M, Timmins, P, Burmeister, W.P, Ruigrok, R.W, Marion, D. | | 登録日 | 2003-10-06 | | 公開日 | 2004-03-09 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and dynamics of the nucleocapsid-binding domain of the Sendai virus phosphoprotein in solution
Virology, 319, 2004
|
|
2C42
 
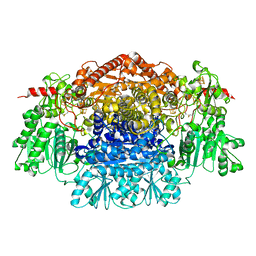 | | Crystal Structure Of Pyruvate-Ferredoxin Oxidoreductase From Desulfovibrio africanus | | 分子名称: | CALCIUM ION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | 著者 | Cavazza, C, Contreras-Martel, C, Pieulle, L, Chabriere, E, Hatchikian, E.C, Fontecilla-Camps, J.C. | | 登録日 | 2005-10-14 | | 公開日 | 2006-12-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Flexibility of Thiamine Diphosphate Revealed by Kinetic Crystallographic Studies of the Reaction of Pyruvate-Ferredoxin Oxidoreductase with Pyruvate.
Structure, 14, 2006
|
|
2BNR
 
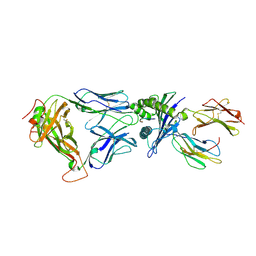 | | Structural and kinetic basis for heightened immunogenicity of T cell vaccines | | 分子名称: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, SYNTHETIC PEPTIDE, ... | | 著者 | Chen, J.-L, Stewart-Jones, G, Bossi, G, Lissin, N.M, Wooldridge, L, Choi, E.M.L, Held, G, Dunbar, P.R, Esnouf, R.M, Sami, M, Boultier, J.M, Rizkallah, P.J, Renner, C, Sewell, A, van der Merwe, P.A, Jackobsen, B.K, Griffiths, G, Jones, E.Y, Cerundolo, V. | | 登録日 | 2005-03-31 | | 公開日 | 2005-05-23 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and Kinetic Basis for Heightened Immunogenicity of T Cell Vaccines
J.Exp.Med., 201, 2005
|
|
4XKC
 
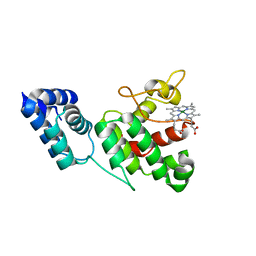 | |
4Q3H
 
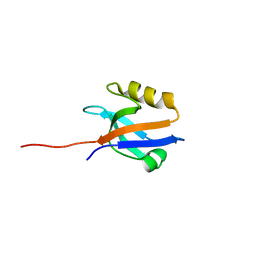 | | The crystal structure of NHERF1 PDZ2 CXCR2 complex revealed by the NHERF1 CXCR2 chimeric protein | | 分子名称: | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | 著者 | Holcomb, J, Jiang, Y, Trescott, L, Lu, G, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | 登録日 | 2014-04-11 | | 公開日 | 2014-05-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.443 Å) | | 主引用文献 | Crystal structure of the NHERF1 PDZ2 domain in complex with the chemokine receptor CXCR2 reveals probable modes of PDZ2 dimerization.
Biochem.Biophys.Res.Commun., 448, 2014
|
|
7TBK
 
 | | Composite structure of the dilated human nuclear pore complex (NPC) symmetric core generated with a 37A in situ cryo-ET map of CD4+ T cell NPC | | 分子名称: | NUP107 CTD, NUP107 NTD, NUP133, ... | | 著者 | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-15 | | 最終更新日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (37 Å) | | 主引用文献 | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7TBI
 
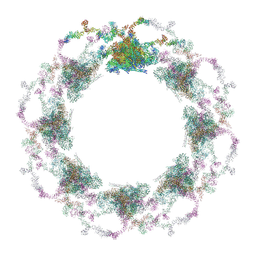 | | Composite structure of the S. cerevisiae nuclear pore complex (NPC) | | 分子名称: | Dyn2, Nic96 R1, Nic96 R2, ... | | 著者 | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-15 | | 最終更新日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (25 Å) | | 主引用文献 | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7TBJ
 
 | | Composite structure of the human nuclear pore complex (NPC) symmetric core generated with a 12A cryo-ET map of the purified HeLa cell NPC | | 分子名称: | NUP107 CTD, NUP107 NTD, NUP133, ... | | 著者 | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-22 | | 最終更新日 | 2022-06-29 | | 実験手法 | ELECTRON MICROSCOPY (23 Å) | | 主引用文献 | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
2BGQ
 
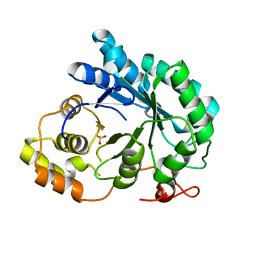 | | apo aldose reductase from barley | | 分子名称: | ALDOSE REDUCTASE, SULFATE ION | | 著者 | Olsen, J.G, Pedersen, L, Christensen, C.L, Olsen, O, Henriksen, A. | | 登録日 | 2005-01-04 | | 公開日 | 2006-06-22 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Barley Aldose Reductase: Structure, Cofactor Binding, and Substrate Recognition in the Aldo/Keto Reductase 4C Family.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
7B5A
 
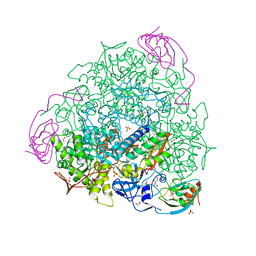 | | X-ray crystal structure of Sporosarcina pasteurii urease inhibited by Ag(PEt3)2NO3 determined at 1.97 Angstroms | | 分子名称: | 1,2-ETHANEDIOL, NICKEL (II) ION, OXYGEN ATOM, ... | | 著者 | Mazzei, L, Cianci, M, Ciurli, S. | | 登録日 | 2020-12-03 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Kinetic and structural analysis of the inactivation of urease by mixed-ligand phosphine halide Ag(I) complexes.
J.Inorg.Biochem., 218, 2021
|
|
7B58
 
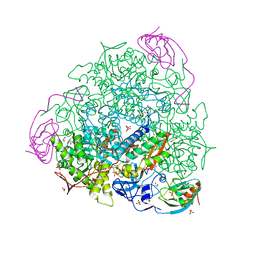 | | X-ray crystal structure of Sporosarcina pasteurii urease inhibited by Ag(PEt3)Cl determined at 1.72 Angstroms | | 分子名称: | 1,2-ETHANEDIOL, NICKEL (II) ION, OXYGEN ATOM, ... | | 著者 | Mazzei, L, Cianci, M, Ciurli, S. | | 登録日 | 2020-12-03 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Kinetic and structural analysis of the inactivation of urease by mixed-ligand phosphine halide Ag(I) complexes.
J.Inorg.Biochem., 218, 2021
|
|
7B59
 
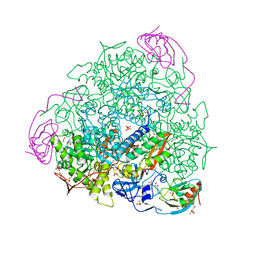 | | X-ray crystal structure of Sporosarcina pasteurii urease inhibited by Ag(PEt3)Br determined at 1.63 Angstroms | | 分子名称: | 1,2-ETHANEDIOL, NICKEL (II) ION, OXYGEN ATOM, ... | | 著者 | Mazzei, L, Cianci, M, Ciurli, S. | | 登録日 | 2020-12-03 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Kinetic and structural analysis of the inactivation of urease by mixed-ligand phosphine halide Ag(I) complexes.
J.Inorg.Biochem., 218, 2021
|
|
4Q7F
 
 | | 1.98 Angstrom Crystal Structure of Putative 5'-Nucleotidase from Staphylococcus aureus in complex with Adenosine. | | 分子名称: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, 5' nucleotidase family protein, MAGNESIUM ION, ... | | 著者 | Minasov, G, Nocadello, S, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-04-24 | | 公開日 | 2014-05-07 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | 1.98 Angstrom Crystal Structure of Putative 5'-Nucleotidase from Staphylococcus aureus in complex with Adenosine.
TO BE PUBLISHED
|
|
4Y1B
 
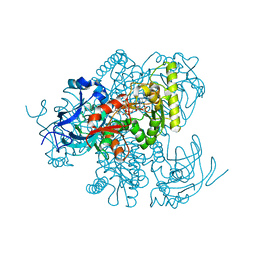 | |
4Y3B
 
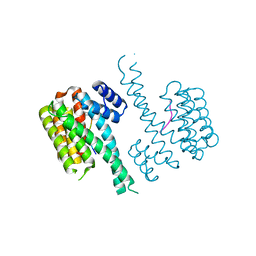 | | Crystal structure of C-terminal modified Tau peptide-hybrid 201D with 14-3-3sigma | | 分子名称: | (2S)-2-(2-methoxyethyl)pyrrolidine, 14-3-3 protein sigma, ARG-THR-PRO-SEP-LEU-PRO-THR-[H][C@@]1(C(C2=CC=CC=C2)C3=CC=CC=C3)CCCN1C | | 著者 | Bartel, M, Milroy, L.G, Brunsveld, L, Ottmann, C. | | 登録日 | 2015-02-10 | | 公開日 | 2015-12-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Stabilizer-Guided Inhibition of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XKB
 
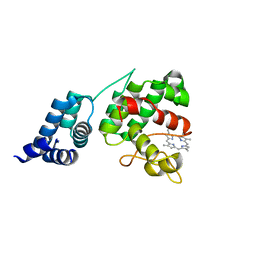 | | Crystal Structure of GENOMES UNCOUPLED 4 (GUN4) in Complex with Deuteroporphyrin IX | | 分子名称: | 3,3'-(3,7,12,17-tetramethylporphyrin-2,18-diyl)dipropanoic acid, Ycf53-like protein | | 著者 | Chen, X, Pu, H, Liu, L. | | 登録日 | 2015-01-11 | | 公開日 | 2015-05-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.501 Å) | | 主引用文献 | Crystal Structures of GUN4 in Complex with Porphyrins.
Mol Plant, 8, 2015
|
|
2DLB
 
 | | X-ray Crystal Structure of Protein yopT from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR412 | | 分子名称: | yopT | | 著者 | Kuzin, A.P, Chen, Y, Seetharaman, J, Ho, C.-K, Cunningham, K, Janjua, H, Conover, K, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-04-18 | | 公開日 | 2006-04-25 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | X-ray structure of hypothetical protein from Bacillus subtilis O34498 at the resolution of 1.2A. NESG target SR412
To be published
|
|
4QF2
 
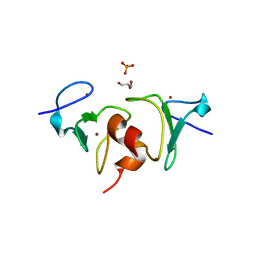 | | Crystal structure of human BAZ2A PHD zinc finger in the free form | | 分子名称: | Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Tallant, C, Overvoorde, L, Van Molle, I, Chirgadze, D.Y, Ciulli, A. | | 登録日 | 2014-05-19 | | 公開日 | 2014-07-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure, 23, 2015
|
|
4QGU
 
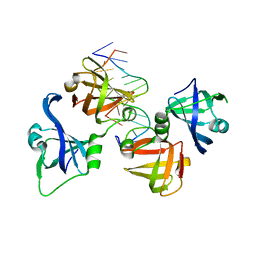 | | protein domain complex with ssDNA | | 分子名称: | DNA (5'-D(P*AP*GP*GP*CP*CP*GP*GP*CP*GP*TP*GP*A)-3'), Gamma-interferon-inducible protein 16 | | 著者 | Ni, X, Ru, H, Zhao, L, Shaw, N, Ding, W, Songying, O, Liu, Z.-J. | | 登録日 | 2014-05-25 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.545 Å) | | 主引用文献 | New insights into the structural basis of DNA recognition by HINa and HINb domains of IFI16.
J Mol Cell Biol, 8, 2016
|
|
4QDZ
 
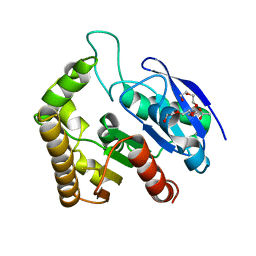 | | Crystal structure of Antigen 85C-E228Q mutant | | 分子名称: | ACETATE ION, Diacylglycerol acyltransferase/mycolyltransferase Ag85C, HEXAETHYLENE GLYCOL | | 著者 | Favrot, L, Lajiness, D.H, Ronning, D.R. | | 登録日 | 2014-05-14 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.885 Å) | | 主引用文献 | Inactivation of the Mycobacterium tuberculosis Antigen 85 Complex by Covalent, Allosteric Inhibitors.
J.Biol.Chem., 289, 2014
|
|
4QE9
 
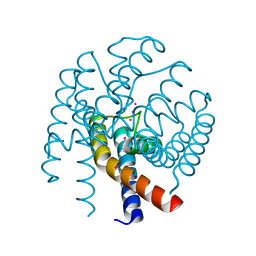 | | Open MthK pore structure soaked in 10 mM Ba2+/100 mM K+ | | 分子名称: | Calcium-gated potassium channel MthK, POTASSIUM ION | | 著者 | Guo, R, Zeng, W, Cui, H, Chen, L, Ye, S. | | 登録日 | 2014-05-15 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Ionic interactions of Ba2+ blockades in the MthK K+ channel
J.Gen.Physiol., 144, 2014
|
|
4QIR
 
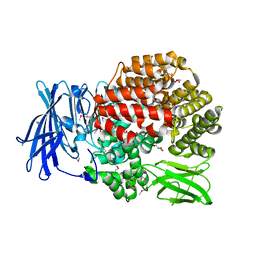 | | Crystal structure of Aminopeptidase N in complex with the phosphinic dipeptide analogue LL-(R,S)-2-(pyridin-3-yl)ethylGlyP[CH2]Phe | | 分子名称: | 3-{[(R)-1-amino-3-(pyridin-3-yl)propyl](hydroxy)phosphoryl}-(S)-2-benzylpropanoic acid, Aminopeptidase N, GLYCEROL, ... | | 著者 | Nocek, B, Joachimiak, A, Berlicki, L, Vassiliou, S, Mucha, A. | | 登録日 | 2014-06-01 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | Structure-guided, single-point modifications in the phosphinic dipeptide structure yield highly potent and selective inhibitors of neutral aminopeptidases.
J.Med.Chem., 57, 2014
|
|
4XRR
 
 | | Crystal structure of cals8 from micromonospora echinospora (P294S mutant) | | 分子名称: | CalS8, GLYCEROL | | 著者 | Michalska, K, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-01-21 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural Characterization of CalS8, a TDP-alpha-D-Glucose Dehydrogenase Involved in Calicheamicin Aminodideoxypentose Biosynthesis.
J. Biol. Chem., 290, 2015
|
|
