5UKO
 
 | |
8BGN
 
 | | N,N-diacetylchitobiose deacetylase from Pyrococcus chitonophagus | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Diacetylchitobiose deacetylase, ... | | 著者 | Rypniewski, W, Bejger, M, Biniek-Antosiak, K. | | 登録日 | 2022-10-28 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Structural, Thermodynamic and Enzymatic Characterization of N , N -Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus.
Int J Mol Sci, 23, 2022
|
|
8BGO
 
 | | N,N-diacetylchitobiose deacetylase from Pyrococcus chitonophagus with substrate N,N-diacetylchitobiose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Diacetylchitobiose deacetylase, ZINC ION | | 著者 | Rypniewski, W, Bejger, M, Biniek-Antosiak, K. | | 登録日 | 2022-10-28 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.08 Å) | | 主引用文献 | Structural, Thermodynamic and Enzymatic Characterization of N , N -Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus.
Int J Mol Sci, 23, 2022
|
|
8BGP
 
 | | N,N-diacetylchitobiose deacetylase from Pyrococcus chitonophagus anomalous data | | 分子名称: | Diacetylchitobiose deacetylase, ZINC ION | | 著者 | Rypniewski, W, Biniek-Antosiak, K, Bejger, M. | | 登録日 | 2022-10-28 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Structural, Thermodynamic and Enzymatic Characterization of N , N -Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus.
Int J Mol Sci, 23, 2022
|
|
2ULL
 
 | |
5LXL
 
 | | NMR structure of the N-terminal domain of the Bacteriophage T5 decoration protein pb10 | | 分子名称: | Decoration protein | | 著者 | Vernhes, E, Gilquin, B, Cuniasse, P, Boulanger, P, Zinn-Justin, S. | | 登録日 | 2016-09-22 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid.
Sci Rep, 7, 2017
|
|
6VVJ
 
 | | Cap1G-TPUA | | 分子名称: | RNA (130-MER) | | 著者 | Summers, M.F, Brown, J.D. | | 登録日 | 2020-02-18 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for transcriptional start site control of HIV-1 RNA fate.
Science, 368, 2020
|
|
6VU1
 
 | |
8A46
 
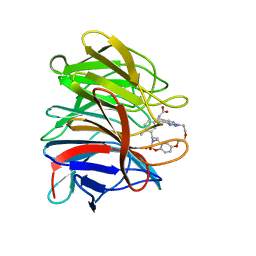 | | Crystal structure of the human Kelch domain of Keap1 in complex with compound S217879 | | 分子名称: | 2-[(1S,2R,8S)-2,4,32-trimethyl-28,28-bis(oxidanylidene)-19,22,27-trioxa-28$l^{6}-thia-1,14,15,16-tetrazahexacyclo[21.5.3.1^{3,7}.1^{9,13}.0^{12,16}.0^{26,30}]tritriaconta-3(33),4,6,9(32),10,12,14,23,25,30-decaen-8-yl]ethanoic acid, Kelch-like ECH-associated protein 1 | | 著者 | Weber, C, Vuillard, L, Delerive, P, Miallau, L. | | 登録日 | 2022-06-10 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.323 Å) | | 主引用文献 | Selective disruption of NRF2-KEAP1 interaction leads to NASH resolution and reduction of liver fibrosis in mice.
JHEP Rep, 5, 2023
|
|
6DE7
 
 | |
5UKQ
 
 | |
4GUM
 
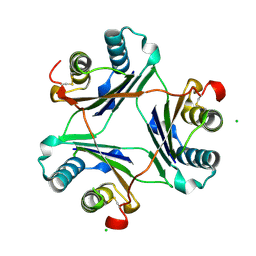 | | Cystal structure of locked-trimer of human MIF | | 分子名称: | CHLORIDE ION, Macrophage migration inhibitory factor | | 著者 | Fan, C, Lolis, E. | | 登録日 | 2012-08-29 | | 公開日 | 2013-07-03 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | MIF intersubunit disulfide mutant antagonist supports activation of CD74 by endogenous MIF trimer at physiologic concentrations.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1SGM
 
 | |
1GBD
 
 | |
1G9C
 
 | |
2H5D
 
 | | 0.9A resolution crystal structure of alpha-lytic protease complexed with a transition state analogue, MeOSuc-Ala-Ala-Pro-Val boronic acid | | 分子名称: | ALPHA-LYTIC PROTEASE, GLYCEROL, MEOSUC-ALA-ALA-PRO-ALA BORONIC ACID INHIBITOR, ... | | 著者 | Fuhrmann, C.N, Agard, D.A. | | 登録日 | 2006-05-25 | | 公開日 | 2006-09-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Subangstrom crystallography reveals that short ionic hydrogen bonds, and not a His-Asp low-barrier hydrogen bond, stabilize the transition state in serine protease catalysis
J.Am.Chem.Soc., 128, 2006
|
|
1GBI
 
 | |
1GBL
 
 | |
1GBJ
 
 | | ALPHA-LYTIC PROTEASE WITH MET 190 REPLACED BY ALA | | 分子名称: | ALPHA-LYTIC PROTEASE, SULFATE ION | | 著者 | Mace, J.E, Agard, D.A. | | 登録日 | 1995-09-06 | | 公開日 | 1996-01-29 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Kinetic and structural characterization of mutations of glycine 216 in alpha-lytic protease: a new target for engineering substrate specificity.
J.Mol.Biol., 254, 1995
|
|
1GBC
 
 | |
1GBE
 
 | |
1GBH
 
 | |
6BXI
 
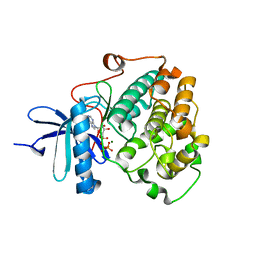 | | X-ray crystal structure of NDR1 kinase domain | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase 38 | | 著者 | Xiong, S, Sicheri, F. | | 登録日 | 2017-12-18 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Basis for Auto-Inhibition of the NDR1 Kinase Domain by an Atypically Long Activation Segment.
Structure, 26, 2018
|
|
1G9A
 
 | |
1GBA
 
 | |
