5HDV
 
 | |
5ZO0
 
 | |
5ER7
 
 | | Connexin-26 Bound to Calcium | | 分子名称: | CALCIUM ION, Gap junction beta-2 protein | | 著者 | Purdy, M.D, Bennett, B.C, Baker, K.A, Yeager, M.J. | | 登録日 | 2015-11-13 | | 公開日 | 2016-01-27 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (3.286 Å) | | 主引用文献 | An electrostatic mechanism for Ca(2+)-mediated regulation of gap junction channels.
Nat Commun, 7, 2016
|
|
6WIQ
 
 | | Crystal structure of the co-factor complex of NSP7 and the C-terminal domain of NSP8 from SARS CoV-2 | | 分子名称: | Non-structural protein 7, Non-structural protein 8 | | 著者 | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-10 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
6WQD
 
 | | The 1.95 A Crystal Structure of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS-CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, Non-structural protein 7, Non-structural protein 8 | | 著者 | Kim, Y, Wilamowski, M, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-28 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
5ERA
 
 | |
8UUZ
 
 | | Campylobacter jejuni CosR apo form | | 分子名称: | DNA-binding response regulator | | 著者 | Zhang, Z. | | 登録日 | 2023-11-02 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.77 Å) | | 主引用文献 | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
8UVX
 
 | | CosR DNA bound form I | | 分子名称: | DNA (5'-D(*AP*TP*AP*TP*CP*TP*TP*AP*AP*TP*TP*TP*TP*GP*GP*TP*TP*AP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*TP*AP*AP*CP*CP*AP*AP*AP*AP*TP*TP*AP*AP*GP*AP*TP*AP*T)-3'), DNA-binding response regulator | | 著者 | Zhang, Z. | | 登録日 | 2023-11-05 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
8UVK
 
 | | CosR DNA bound form II | | 分子名称: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*AP*AP*TP*TP*AP*AP*GP*AP*TP*AP*TP*TP*AP*TP*TP*AP*AP*CP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*GP*TP*TP*AP*AP*TP*AP*AP*TP*AP*TP*CP*TP*TP*AP*AP*TP*T)-3'), ... | | 著者 | Zhang, Z. | | 登録日 | 2023-11-03 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
6XIP
 
 | | The 1.5 A Crystal Structure of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, Non-structural protein 7, Non-structural protein 8 | | 著者 | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-06-20 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
7KJS
 
 | | Crystal structure of CDK2/cyclin E in complex with PF-06873600 | | 分子名称: | 6-(difluoromethyl)-8-[(1R,2R)-2-hydroxy-2-methylcyclopentyl]-2-{[1-(methylsulfonyl)piperidin-4-yl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | 著者 | McTigue, M.A, He, Y, Ferre, R.A. | | 登録日 | 2020-10-26 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.187 Å) | | 主引用文献 | Discovery of PF-06873600, a CDK2/4/6 Inhibitor for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
7D0C
 
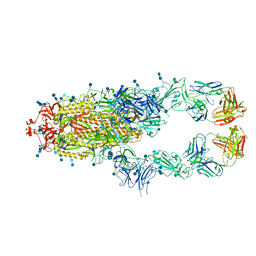 | | S protein of SARS-CoV-2 in complex bound with P5A-3A1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of P5A-3A1, ... | | 著者 | Yan, R.H, Wang, R.K, Yu, J.F, Zhang, Y.Y, Liu, N, Wang, H.W, Wang, X.Q, Zhang, L.Q, Zhou, Q. | | 登録日 | 2020-09-09 | | 公開日 | 2021-03-10 | | 最終更新日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7D0B
 
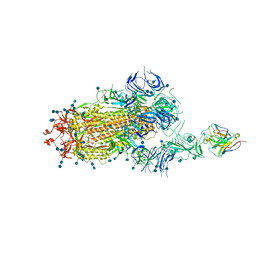 | | S protein of SARS-CoV-2 in complex bound with P5A-3C12_1B | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of P5A-3C12, ... | | 著者 | Yan, R.H, Wang, R.K, Ju, B, Yu, J.F, Zhang, Y.Y, Liu, N, Wang, H.W, Wang, X.Q, Zhang, L.Q, Zhou, Q. | | 登録日 | 2020-09-09 | | 公開日 | 2021-03-10 | | 最終更新日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7D0D
 
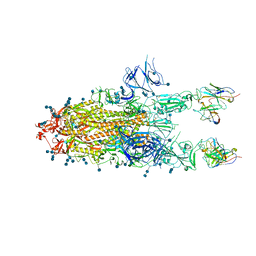 | | S protein of SARS-CoV-2 in complex bound with P5A-3C12_2B | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of P5A-3C12, ... | | 著者 | Yan, R.H, Wang, R.K, Ju, B, Yu, J.F, Zhang, Y.Y, Liu, N, Wang, H.W, Wang, X.Q, Zhang, L.Q, Zhou, Q. | | 登録日 | 2020-09-09 | | 公開日 | 2021-03-10 | | 最終更新日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
5XVA
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH216 | | 分子名称: | ETHANOL, Serine/threonine-protein kinase PAK 4, [6-chloranyl-4-[(5-methyl-1H-pyrazol-3-yl)amino]quinazolin-2-yl]-[(3R)-3-methylpiperazin-1-yl]methanone | | 著者 | Zhao, F, Li, H. | | 登録日 | 2017-06-27 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.847 Å) | | 主引用文献 | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5XVG
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH226 | | 分子名称: | 1,2-ETHANEDIOL, ETHANOL, Serine/threonine-protein kinase PAK 4, ... | | 著者 | Zhao, F, Li, H. | | 登録日 | 2017-06-27 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
7FAE
 
 | | S protein of SARS-CoV-2 in complex bound with P36-5D2(state2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P36-5D2 heavy chain, ... | | 著者 | Zhang, L, Wang, X, Shan, S, Zhang, S. | | 登録日 | 2021-07-06 | | 公開日 | 2021-12-22 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | A Potent and Protective Human Neutralizing Antibody Against SARS-CoV-2 Variants.
Front Immunol, 12, 2021
|
|
7FAF
 
 | | S protein of SARS-CoV-2 in complex bound with P36-5D2 (state1) | | 分子名称: | P36-5D2 heavy chain, P36-5D2 light chain, Spike glycoprotein | | 著者 | Zhang, L, Wang, X, Zhang, S, Shan, S. | | 登録日 | 2021-07-06 | | 公開日 | 2021-12-22 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.69 Å) | | 主引用文献 | A Potent and Protective Human Neutralizing Antibody Against SARS-CoV-2 Variants.
Front Immunol, 12, 2021
|
|
5XXI
 
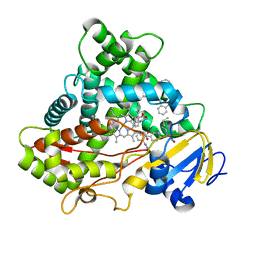 | | Crystal structure of CYP2C9 in complex with multiple losartan molecules | | 分子名称: | Cytochrome P450 2C9, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Maekawa, K, Adachi, M, Shah, M.B. | | 登録日 | 2017-07-04 | | 公開日 | 2017-10-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Basis of Single-Nucleotide Polymorphisms in Cytochrome P450 2C9
Biochemistry, 56, 2017
|
|
7CJF
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | 著者 | Guo, Y, Li, X, Zhang, G, Fu, D, Schweizer, L, Zhang, H, Rao, Z. | | 登録日 | 2020-07-10 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.108 Å) | | 主引用文献 | A SARS-CoV-2 neutralizing antibody with extensive Spike binding coverage and modified for optimal therapeutic outcomes.
Nat Commun, 12, 2021
|
|
5X23
 
 | | Crystal structure of CYP2C9 genetic variant A477T (*30) in complex with multiple losartan molecules | | 分子名称: | Cytochrome P450 2C9, PHOSPHATE ION, POTASSIUM ION, ... | | 著者 | Maekawa, K, Adachi, M, Shah, M.B. | | 登録日 | 2017-01-30 | | 公開日 | 2017-10-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis of Single-Nucleotide Polymorphisms in Cytochrome P450 2C9
Biochemistry, 56, 2017
|
|
5YMU
 
 | |
5WBG
 
 | | Crystal Structure of human Cytochrome P450 2B6 (Y226H/K262R) in complex with an analog of a drug Efavirenz | | 分子名称: | (2R,4S)-6-chloro-4-(cyclopropylethynyl)-2-methyl-4-(trifluoromethyl)-1,4-dihydro-2H-3,1-benzoxazine, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, 5-cyclohexylpentan-1-ol, ... | | 著者 | Shah, M.B, Halpert, J.R. | | 登録日 | 2017-06-29 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Crystal Structure of CYP2B6 in Complex with an Efavirenz Analog.
Int J Mol Sci, 19, 2018
|
|
5WQN
 
 | |
5YSX
 
 | |
