7XX2
 
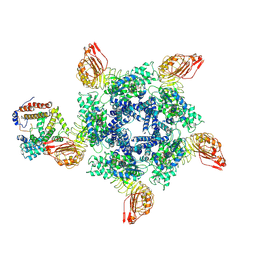 | | Cryo-EM structure of Sr35 resistosome induced by AvrSr35 R381A | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, AvrSr35, CNL9 | | 著者 | Ouyang, S.Y, Zhao, Y.B, Li, Z.K, Liu, M.X. | | 登録日 | 2022-05-28 | | 公開日 | 2022-11-02 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Pathogen effector AvrSr35 triggers Sr35 resistosome assembly via a direct recognition mechanism.
Sci Adv, 8, 2022
|
|
8JYD
 
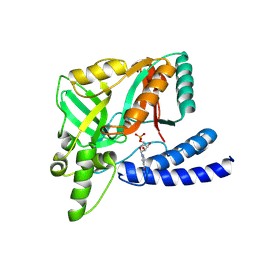 | |
6KCW
 
 | | Structure of alginate lyase Aly36B | | 分子名称: | Alginate lyase, CALCIUM ION, PHOSPHATE ION | | 著者 | Dong, F, Chen, X.L, Zhang, Y.Z. | | 登録日 | 2019-06-29 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Alginate Lyase Aly36B is a New Bacterial Member of the Polysaccharide Lyase Family 36 and Catalyzes by a Novel Mechanism With Lysine as Both the Catalytic Base and Catalytic Acid.
J.Mol.Biol., 431, 2019
|
|
6K7Y
 
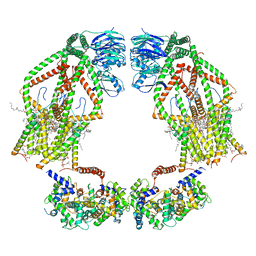 | | Intact human mitochondrial calcium uniporter complex with MICU1/MICU2 subunits | | 分子名称: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, CALCIUM ION, CARDIOLIPIN, ... | | 著者 | Zhuo, W, Zhou, H, Yang, M. | | 登録日 | 2019-06-10 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure of intact human MCU supercomplex with the auxiliary MICU subunits.
Protein Cell, 12, 2021
|
|
6KL9
 
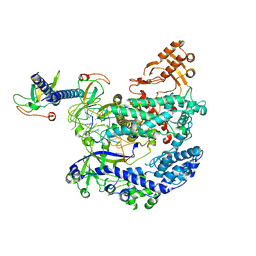 | | Structure of LbCas12a-crRNA complex bound to AcrVA4 (form A complex) | | 分子名称: | AcrVA4, LbCas12a, MAGNESIUM ION, ... | | 著者 | Peng, R, Li, Z, Xu, Y, He, S, Peng, Q, Shi, Y, Gao, G.F. | | 登録日 | 2019-07-30 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Structural insight into multistage inhibition of CRISPR-Cas12a by AcrVA4.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K7X
 
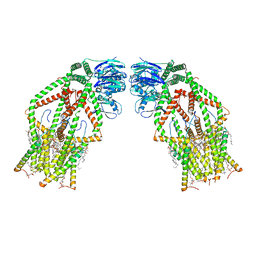 | | Human MCU-EMRE complex | | 分子名称: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, CALCIUM ION, CARDIOLIPIN, ... | | 著者 | Zhuo, W, Zhou, H, Yang, M. | | 登録日 | 2019-06-10 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Structure of intact human MCU supercomplex with the auxiliary MICU subunits.
Protein Cell, 12, 2021
|
|
6KCV
 
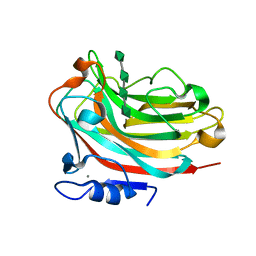 | |
1J4K
 
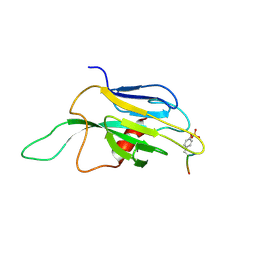 | |
6KLB
 
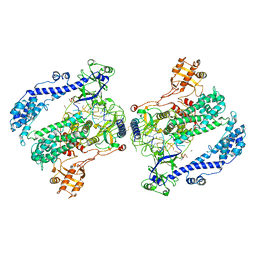 | | Structure of LbCas12a-crRNA complex bound to AcrVA4 (form B complex) | | 分子名称: | AcrVA4, LbCas12a, MAGNESIUM ION, ... | | 著者 | Peng, R, Li, Z, Xu, Y, He, S, Peng, Q, Shi, Y, Gao, G.F. | | 登録日 | 2019-07-30 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural insight into multistage inhibition of CRISPR-Cas12a by AcrVA4.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1K2M
 
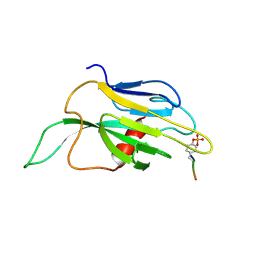 | |
6LA5
 
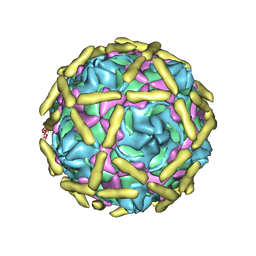 | |
6LB1
 
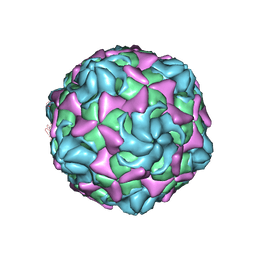 | |
6LA7
 
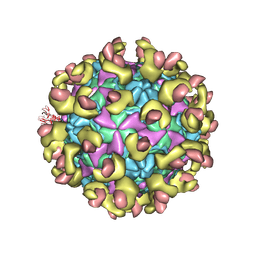 | |
7WID
 
 | | Crystal structure of Staphylococcus aureus ClpP in complex with ZG180 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, (6S,9aS)-6-[(2S)-butan-2-yl]-8-(naphthalen-1-ylmethyl)-4,7-bis(oxidanylidene)-N-[4,4,4-tris(fluoranyl)butyl]-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, ... | | 著者 | Wei, B.Y, Gan, J.H, Yang, C.-G. | | 登録日 | 2022-01-03 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Anti-infective therapy using species-specific activators of Staphylococcus aureus ClpP.
Nat Commun, 13, 2022
|
|
7W0S
 
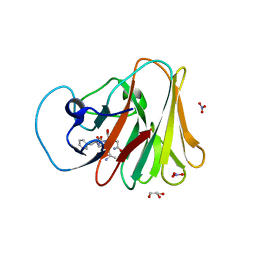 | | TRIM7 in complex with C-terminal peptide of 2C | | 分子名称: | DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase TRIM7, GLYCEROL, ... | | 著者 | Zhang, H, Liang, X, Li, X.Z. | | 登録日 | 2021-11-18 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | A C-terminal glutamine recognition mechanism revealed by E3 ligase TRIM7 structures.
Nat.Chem.Biol., 18, 2022
|
|
7W0Q
 
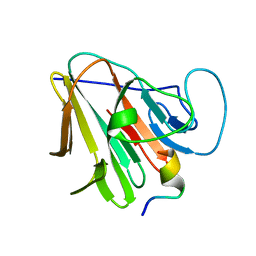 | |
7W0T
 
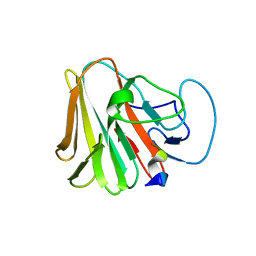 | |
7WGS
 
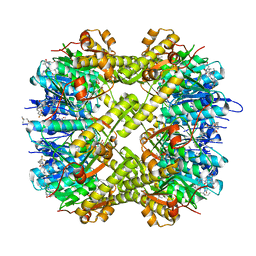 | | Structure of ClpP from Staphylococcus aureus in complex with (S)-ZG197 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, (6S,9aS)-6-[(2R)-butan-2-yl]-8-[(1S)-1-naphthalen-1-ylethyl]-4,7-bis(oxidanylidene)-N-[4,4,4-tris(fluoranyl)butyl]-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit | | 著者 | Yang, C.-G, Gan, J.H. | | 登録日 | 2021-12-28 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Anti-infective therapy using species-specific activators of Staphylococcus aureus ClpP.
Nat Commun, 13, 2022
|
|
7WH5
 
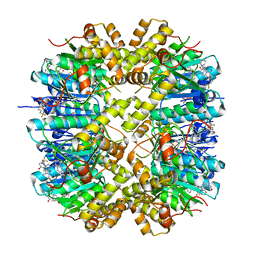 | | Crystal structure of human ClpP in complex with ZG180 | | 分子名称: | (6S,9aS)-6-[(2S)-butan-2-yl]-8-(naphthalen-1-ylmethyl)-4,7-bis(oxidanylidene)-N-[4,4,4-tris(fluoranyl)butyl]-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, mitochondrial, ... | | 著者 | Wei, B.Y, Gan, J.H, Yang, C.-G. | | 登録日 | 2021-12-29 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Anti-infective therapy using species-specific activators of Staphylococcus aureus ClpP.
Nat Commun, 13, 2022
|
|
8YNZ
 
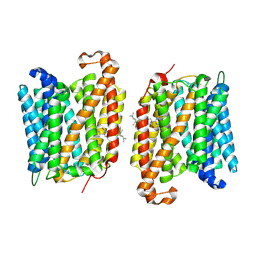 | | The structure of EfpA_BRD-8000.3 complex | | 分子名称: | (1S,3S)-N-[6-bromo-5-(pyrimidin-2-yl)pyridin-2-yl]-2,2-dimethyl-3-(2-methylprop-1-en-1-yl)cyclopropane-1-carboxamide, Uncharacterized MFS-type transporter EfpA | | 著者 | Li, D.L, Sun, J.Q. | | 登録日 | 2024-03-12 | | 公開日 | 2024-12-04 | | 実験手法 | ELECTRON MICROSCOPY (3.41 Å) | | 主引用文献 | Structure and function of Mycobacterium tuberculosis EfpA as a lipid transporter and its inhibition by BRD-8000.3.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7WOT
 
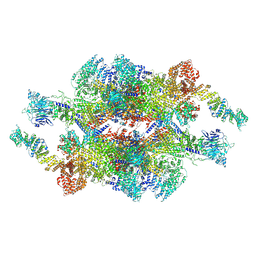 | | Cryo-EM structure of the inner ring monomer of the Saccharomyces cerevisiae nuclear pore complex | | 分子名称: | Nucleoporin NIC96, Nucleoporin NSP1, Nucleoporin NUP157, ... | | 著者 | Li, Z.Q, Chen, S.J.B, Zhao, L, Sui, S.F. | | 登録日 | 2022-01-22 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.73 Å) | | 主引用文献 | Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex.
Cell Res., 32, 2022
|
|
7WOO
 
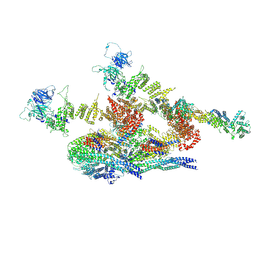 | | Cryo-EM structure of the inner ring protomer of the Saccharomyces cerevisiae nuclear pore complex | | 分子名称: | Nucleoporin NIC96, Nucleoporin NSP1, Nucleoporin NUP157, ... | | 著者 | Li, Z.Q, Chen, S.J.B, Zhao, L, Sui, S.F. | | 登録日 | 2022-01-22 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.71 Å) | | 主引用文献 | Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex.
Cell Res., 32, 2022
|
|
7XDS
 
 | |
7XE0
 
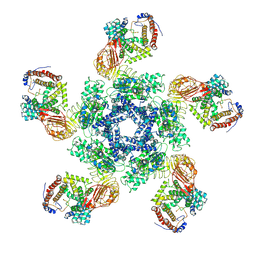 | | Cryo-EM structure of plant NLR Sr35 resistosome | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, AvrSr35, Sr35 | | 著者 | Ouyang, S.Y, Zhao, Y.B, Li, Z.K, Liu, M.X. | | 登録日 | 2022-03-29 | | 公開日 | 2022-09-28 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | Pathogen effector AvrSr35 triggers Sr35 resistosome assembly via a direct recognition mechanism.
Sci Adv, 8, 2022
|
|
7X6Z
 
 | |
