5UCW
 
 | |
6E1C
 
 | |
4D3O
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 6-(3-(2-(1H-Pyrrolo(2,3-b)pyridin-6-yl)ethyl)-5-(aminomethyl) phenethyl)-4-methylpyridin-2-amine | | 分子名称: | 6-(2-{3-(aminomethyl)-5-[2-(1H-pyrrolo[2,3-b]pyridin-6-yl)ethyl]phenyl}ethyl)-4-methylpyridin-2-amine, CHLORIDE ION, GLYCEROL, ... | | 著者 | Holden, J.K, Poulos, T.L. | | 登録日 | 2014-10-23 | | 公開日 | 2015-01-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-Based Design of Bacterial Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 58, 2015
|
|
4D3U
 
 | |
4D3T
 
 | |
4D3J
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 6,6'-(2,2'-(5-amino-1,3-phenylene)bis(ethane-2,1-diyl))bis(4- methylpyridin-2-amine) | | 分子名称: | 6,6'-[(5-aminobenzene-1,3-diyl)diethane-2,1-diyl]bis(4-methylpyridin-2-amine), CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Holden, J.K, Poulos, T.L. | | 登録日 | 2014-10-22 | | 公開日 | 2015-01-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Structure-Based Design of Bacterial Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 58, 2015
|
|
4D3N
 
 | |
4D3I
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 6,6'-((5-(aminomethyl)-1,3-phenylene)bis(ethane-2,1-diyl))bis(4- methylpyridin-2-amine) | | 分子名称: | 6,6'-{[5-(aminomethyl)benzene-1,3-diyl]diethane-2,1-diyl}bis(4-methylpyridin-2-amine), GLYCEROL, N-PROPANOL, ... | | 著者 | Holden, J.K, Poulos, T.L. | | 登録日 | 2014-10-22 | | 公開日 | 2015-01-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structure-Based Design of Bacterial Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 58, 2015
|
|
4D3M
 
 | |
4D3V
 
 | |
4D3K
 
 | | Structure of Bacillus subtilis nitric oxide synthase in complex with 6,6'-((5-(3-aminopropyl)-1,3-phenylene)bis(ethane-2,1-diyl))bis(4- methylpyridin-2-amine) | | 分子名称: | 6,6'-{[5-(3-aminopropyl)benzene-1,3-diyl]diethane-2,1-diyl}bis(4-methylpyridin-2-amine), CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Holden, J.K, Poulos, T.L. | | 登録日 | 2014-10-22 | | 公開日 | 2015-01-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.017 Å) | | 主引用文献 | Structure-Based Design of Bacterial Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 58, 2015
|
|
8ETR
 
 | | CryoEM Structure of NLRP3 NACHT domain in complex with G2394 | | 分子名称: | (6S,8R)-N-[(1,2,3,5,6,7-hexahydro-s-indacen-4-yl)carbamoyl]-6-(methylamino)-6,7-dihydro-5H-pyrazolo[5,1-b][1,3]oxazine-3-sulfonamide, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Murray, J.M, Johnson, M.C. | | 登録日 | 2022-10-17 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Overcoming Preclinical Safety Obstacles to Discover ( S )- N -((1,2,3,5,6,7-Hexahydro- s -indacen-4-yl)carbamoyl)-6-(methylamino)-6,7-dihydro-5 H -pyrazolo[5,1- b ][1,3]oxazine-3-sulfonamide (GDC-2394): A Potent and Selective NLRP3 Inhibitor.
J.Med.Chem., 65, 2022
|
|
6JFK
 
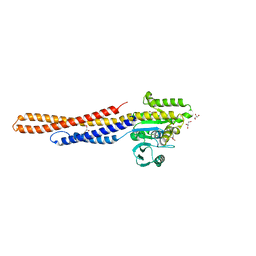 | | GDP bound Mitofusin2 (MFN2) | | 分子名称: | CITRIC ACID, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Li, Y.J, Cao, Y.L, Feng, J.X, Qi, Y.B, Meng, S.X, Yang, J.F, Zhong, Y.T, Kang, S.S, Chen, X.X, Lan, L, Luo, L, Yu, B, Chen, S.D, Chan, D.C, Hu, J.J, Gao, S. | | 登録日 | 2019-02-10 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.997 Å) | | 主引用文献 | Structural insights of human mitofusin-2 into mitochondrial fusion and CMT2A onset.
Nat Commun, 10, 2019
|
|
6JFL
 
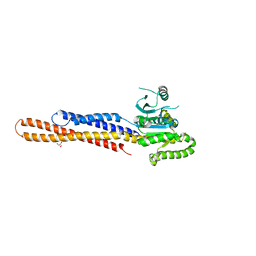 | | Nucleotide-free Mitofusin2 (MFN2) | | 分子名称: | CALCIUM ION, GLYCEROL, Mitofusin-2,cDNA FLJ57997, ... | | 著者 | Li, Y.J, Cao, Y.L, Feng, J.X, Qi, Y.B, Meng, S.X, Yang, J.F, Zhong, Y.T, Kang, S.S, Chen, X.X, Lan, L, Luo, L, Yu, B, Chen, S.D, Chan, D.C, Hu, J.J, Gao, S. | | 登録日 | 2019-02-10 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.806 Å) | | 主引用文献 | Structural insights of human mitofusin-2 into mitochondrial fusion and CMT2A onset.
Nat Commun, 10, 2019
|
|
6JFM
 
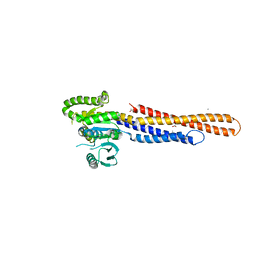 | | Mitofusin2 (MFN2)_T111D | | 分子名称: | ACETATE ION, CALCIUM ION, Mitofusin-2,Mitofusin-2 | | 著者 | Li, Y.J, Cao, Y.L, Feng, J.X, Qi, Y.B, Meng, S.X, Yang, J.F, Zhong, Y.T, Kang, S.S, Chen, X.X, Lan, L, Luo, L, Yu, B, Chen, S.D, Chan, D.C, Hu, J.J, Gao, S. | | 登録日 | 2019-02-10 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structural insights of human mitofusin-2 into mitochondrial fusion and CMT2A onset.
Nat Commun, 10, 2019
|
|
8K5J
 
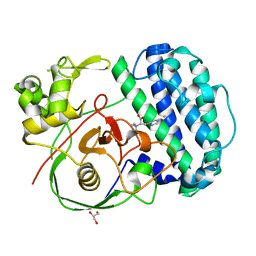 | | The structure of SenA in complex with N,N,N-trimethyl-histidine | | 分子名称: | FE (III) ION, GLYCEROL, N,N,N-trimethyl-histidine, ... | | 著者 | Liu, M, Yang, Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | 登録日 | 2023-07-21 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structural insights into a novel nonheme iron-dependent oxygenase in selenoneine biosynthesis.
Int.J.Biol.Macromol., 256, 2023
|
|
8K5K
 
 | | The structure of SenA | | 分子名称: | FE (III) ION, GLYCEROL, selenoneine synthase SenA | | 著者 | Liu, M, Yang, Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | 登録日 | 2023-07-21 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural insights into a novel nonheme iron-dependent oxygenase in selenoneine biosynthesis.
Int.J.Biol.Macromol., 256, 2023
|
|
8K5I
 
 | | The structure of SenA in complex with N,N,N-trimethyl-histidine and thioglucose | | 分子名称: | 1-thio-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | 著者 | Liu, M, Yang, Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | 登録日 | 2023-07-21 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural insights into a novel nonheme iron-dependent oxygenase in selenoneine biosynthesis.
Int.J.Biol.Macromol., 256, 2023
|
|
2NAE
 
 | |
6WUS
 
 | |
3DEE
 
 | |
2MP0
 
 | | Protein Phosphorylation upon a Fleeting Encounter | | 分子名称: | Glucose-specific phosphotransferase enzyme IIA component, PHOSPHITE ION, Phosphoenolpyruvate-protein phosphotransferase | | 著者 | Xing, Q, Yang, J, Huang, P, Zhang, W, Tang, C. | | 登録日 | 2014-05-08 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Visualizing an ultra-weak protein-protein interaction in phosphorylation signaling.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5BOA
 
 | | Crystal Structure of the Meningitis Pathogen Streptococcus suis adhesion Fhb bound to the disaccharide receptor Gb2 | | 分子名称: | Translation initiation factor 2 (IF-2 GTPase), alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose | | 著者 | Zhang, C, Yu, Y, Yang, M, Jiang, Y. | | 登録日 | 2015-05-27 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.708 Å) | | 主引用文献 | Structural basis of the interaction between the meningitis pathogen Streptococcus suis adhesin Fhb and its human receptor.
Febs Lett., 590, 2016
|
|
3F1Z
 
 | |
5HYN
 
 | | Structure of Human Polycomb Repressive Complex 2 (PRC2) with oncogenic histone H3K27M peptide | | 分子名称: | H3K27M, Histone-lysine N-methyltransferase EZH2, JARID2 K116me3, ... | | 著者 | Zhang, Y, Justin, N, Wilson, J.R, Gamblin, S.J. | | 登録日 | 2016-02-01 | | 公開日 | 2016-05-11 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2.
Nat Commun, 7, 2016
|
|
