7C8K
 
 | | Structural basis for cross-species recognition of COVID-19 virus spike receptor binding domain to bat ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | 著者 | Liu, K.F, Wang, J, Tan, S.G, Niu, S, Wu, L.L, Zhang, Y.F, Pan, X.Q, Meng, Y.M, Chen, Q, Wang, Q.H, Wang, H.W, Qi, J.X, Gao, G.F. | | 登録日 | 2020-06-02 | | 公開日 | 2021-01-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cross-species recognition of SARS-CoV-2 to bat ACE2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8GPX
 
 | | YFV_E_YD73Fab_postfusion | | 分子名称: | Envelope protein, YD73Fab_H, YD73Fab_K | | 著者 | Li, Y, Wu, L, Chai, Y, Qi, J, Yan, J, Gao, G.F. | | 登録日 | 2022-08-27 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | A neutralizing-protective supersite of human monoclonal antibodies for yellow fever virus.
Innovation (N Y), 3, 2022
|
|
8GPU
 
 | | YFV_E_YD6Fab_prefusion | | 分子名称: | Envelope protein, YD6Fab_H, YD6Fab_L | | 著者 | Li, Y, Wu, L, Qi, J, Yan, J, Gao, G.F. | | 登録日 | 2022-08-27 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | A neutralizing-protective supersite of human monoclonal antibodies for yellow fever virus.
Innovation (N Y), 3, 2022
|
|
8GPT
 
 | | YFV_E_YD6scFv_postfusion | | 分子名称: | Envelope protein, YD6_VH, YD6_VL | | 著者 | Li, Y, Wu, L, Qi, J, Yan, J, Gao, G.F. | | 登録日 | 2022-08-27 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | A neutralizing-protective supersite of human monoclonal antibodies for yellow fever virus.
Innovation (N Y), 3, 2022
|
|
8GNJ
 
 | |
8GNI
 
 | |
8GQ5
 
 | |
4RSU
 
 | | Crystal structure of the light and hvem complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | 著者 | Liu, W, Ramagoal, U.A, Himmel, D, Bonanno, J.B, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2014-11-11 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | 分子名称: | 3C-like proteinase | | 著者 | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | 登録日 | 2020-12-24 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7DWO
 
 | |
7DWN
 
 | | Crystal structure of Vibrio fischeri DarR in complex with DNA reveals the transcriptional activation mechanism of LTTR family members | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Predicted DNA-binding transcriptional regulator | | 著者 | Wang, W.W, Wu, H, He, J.H, Yu, F. | | 登録日 | 2021-01-17 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Crystal structure details of Vibrio fischeri DarR and mutant DarR-M202I from LTTR family reveals their activation mechanism.
Int.J.Biol.Macromol., 183, 2021
|
|
8H69
 
 | | Cryo-EM structure of influenza RNA polymerase | | 分子名称: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*UP*AP*AP*AP*CP*UP*CP*CP*UP*GP*CP*UP*UP*UP*UP*GP*CP*U)-3'), ... | | 著者 | Li, H, Wu, Y, Liang, H, Liu, Y. | | 登録日 | 2022-10-16 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | An intermediate state allows influenza polymerase to switch smoothly between transcription and replication cycles.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7EFT
 
 | |
7EO8
 
 | | Crystal structure of SARS coronavirus main protease in complex with an inhibitor Shikonin | | 分子名称: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | 著者 | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | 登録日 | 2021-04-21 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2808516 Å) | | 主引用文献 | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7BW4
 
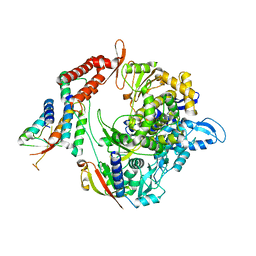 | | Structure of the RNA-dependent RNA polymerase from SARS-CoV-2 | | 分子名称: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | 著者 | Peng, Q, Peng, R, Shi, Y. | | 登録日 | 2020-04-13 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural and Biochemical Characterization of the nsp12-nsp7-nsp8 Core Polymerase Complex from SARS-CoV-2.
Cell Rep, 31, 2020
|
|
8HU4
 
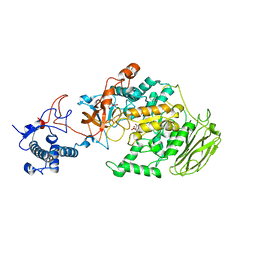 | | Limosilactobacillus reuteri N1 GtfB | | 分子名称: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | 著者 | Dong, J.J, Bai, Y.X. | | 登録日 | 2022-12-22 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Insights into the Structure-Function Relationship of GH70 GtfB alpha-Glucanotransferases from the Crystal Structure and Molecular Dynamic Simulation of a Newly Characterized Limosilactobacillus reuteri N1 GtfB Enzyme.
J.Agric.Food Chem., 72, 2024
|
|
5CNO
 
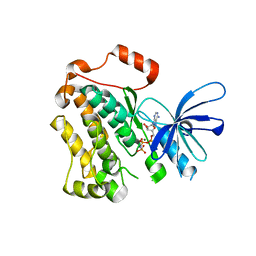 | | Crystal structure of the EGFR kinase domain mutant V924R | | 分子名称: | Epidermal growth factor receptor, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Kovacs, E, Das, R, Mirza, A, Jura, N, Barros, T, Kuriyan, J. | | 登録日 | 2015-07-17 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Analysis of the Role of the C-Terminal Tail in the Regulation of the Epidermal Growth Factor Receptor.
Mol.Cell.Biol., 35, 2015
|
|
6J6M
 
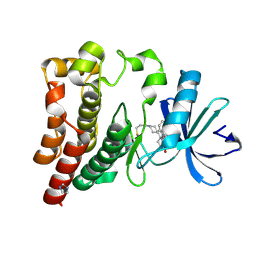 | | Co-crystal structure of BTK kinase domain with Zanubrutinib | | 分子名称: | (7S)-2-(4-phenoxyphenyl)-7-(1-propanoylpiperidin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, IMIDAZOLE, Tyrosine-protein kinase BTK | | 著者 | Zhou, X, Hong, Y. | | 登録日 | 2019-01-15 | | 公開日 | 2019-10-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Discovery of Zanubrutinib (BGB-3111), a Novel, Potent, and Selective Covalent Inhibitor of Bruton's Tyrosine Kinase.
J.Med.Chem., 62, 2019
|
|
8HWT
 
 | |
8HWS
 
 | | The complex structure of Omicron BA.4 RBD with BD604, S309, and S304 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD-604 Fab Heavy chain, BD-604 Fab Light chain, ... | | 著者 | He, Q.W, Xu, Z.P, Xie, Y.F. | | 登録日 | 2023-01-02 | | 公開日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (2.36 Å) | | 主引用文献 | An updated atlas of antibody evasion by SARS-CoV-2 Omicron sub-variants including BQ.1.1 and XBB.
Cell Rep Med, 4, 2023
|
|
8HWK
 
 | | Limosilactobacillus reuteri N1 GtfB-maltohexaose | | 分子名称: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | 著者 | Dong, J.J, Bai, Y.X. | | 登録日 | 2022-12-30 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Insights into the Structure-Function Relationship of GH70 GtfB alpha-Glucanotransferases from the Crystal Structure and Molecular Dynamic Simulation of a Newly Characterized Limosilactobacillus reuteri N1 GtfB Enzyme.
J.Agric.Food Chem., 72, 2024
|
|
8HW3
 
 | | Limosilactobacillus reuteri N1 GtfB-acarbose | | 分子名称: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, GLYCEROL, SODIUM ION, ... | | 著者 | Dong, J.J, Bai, Y.X. | | 登録日 | 2022-12-28 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Insights into the Structure-Function Relationship of GH70 GtfB alpha-Glucanotransferases from the Crystal Structure and Molecular Dynamic Simulation of a Newly Characterized Limosilactobacillus reuteri N1 GtfB Enzyme.
J.Agric.Food Chem., 72, 2024
|
|
7F9I
 
 | | The apo-form structure of EnrR | | 分子名称: | EnrR repressor | | 著者 | Gan, J.H, Wang, Q.Y. | | 登録日 | 2021-07-04 | | 公開日 | 2022-05-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Xenogeneic nucleoid-associated EnrR thwarts H-NS silencing of bacterial virulence with unique DNA binding.
Nucleic Acids Res., 50, 2022
|
|
7F9H
 
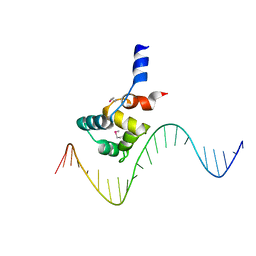 | | complex structure of EnrR-DNA | | 分子名称: | EnrR repressor, target DNA | | 著者 | Gan, J.H, Wang, Q.Y. | | 登録日 | 2021-07-04 | | 公開日 | 2022-05-11 | | 最終更新日 | 2022-05-18 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Xenogeneic nucleoid-associated EnrR thwarts H-NS silencing of bacterial virulence with unique DNA binding.
Nucleic Acids Res., 50, 2022
|
|
8JMT
 
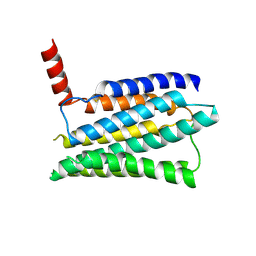 | | Structure of the adhesion GPCR ADGRL3 in the apo state | | 分子名称: | Adhesion G protein-coupled receptor L3,Soluble cytochrome b562 | | 著者 | Tao, Y, Guo, Q, He, B, Zhong, Y. | | 登録日 | 2023-06-05 | | 公開日 | 2023-09-06 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.36 Å) | | 主引用文献 | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
