4FL2
 
 | | Structural and Biophysical Characterization of the Syk Activation Switch | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Tyrosine-protein kinase SYK | | 著者 | Graedler, U, Schwarz, D, Dresing, V, Musil, M, Bomke, J, Frech, M, Jaekel, S, Rysiok, T, Mueller-Pompalla, D, Wegener, A. | | 登録日 | 2012-06-14 | | 公開日 | 2012-11-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Structural and biophysical characterization of the syk activation switch.
J.Mol.Biol., 425, 2013
|
|
8X82
 
 | |
4BQJ
 
 | | structure of HSP90 with an inhibitor bound | | 分子名称: | 5-[2,4-dihydroxy-6-(4-nitrophenoxy)phenyl]-N-ethyl-1,2-oxazole-3-carboxamide, HEAT SHOCK PROTEIN HSP 90-ALPHA | | 著者 | Casale, E, Brasca, M.G, Mantegani, S, Amboldi, N, Bindi, S, Caronni, D, Ceccarelli, W, Colombo, N, DePonti, A, Donati, D, Ermoli, A, Fachin, G, Felder, E.R, Ferguson, R.D, Fiorelli, C, Guanci, M, Isacchi, A, Pesenti, E, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Fogliatto, G. | | 登録日 | 2013-05-30 | | 公開日 | 2013-10-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of Nms-E973 as Novel, Selective and Potent Inhibitor of Heat Shock Protein 90 (Hsp90).
Bioorg.Med.Chem., 21, 2013
|
|
3B8W
 
 | | Crystal structure of Escherichia coli alaine racemase mutant E221P | | 分子名称: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | 登録日 | 2007-11-02 | | 公開日 | 2008-07-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
8X84
 
 | |
2EGH
 
 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase complexed with a magnesium ion, NADPH and fosmidomycin | | 分子名称: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MAGNESIUM ION, ... | | 著者 | Yajima, S, Hara, K, Iino, D, Sasaki, Y, Kuzuyama, T, Seto, H. | | 登録日 | 2007-03-01 | | 公開日 | 2007-06-19 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase in a quaternary complex with a magnesium ion, NADPH and the antimalarial drug fosmidomycin
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2JM1
 
 | |
3BB6
 
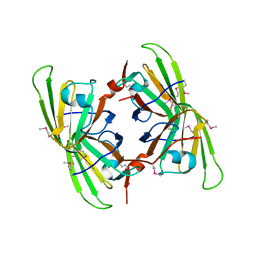 | | Crystal structure of the P64488 protein from E.coli (strain K12). Northeast Structural Genomics Consortium target ER596 | | 分子名称: | Uncharacterized protein yeaR, ZINC ION | | 著者 | Kuzin, A.P, Su, M, Seetharaman, J, Wang, D, Janjua, H, Owens, L, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-11-09 | | 公開日 | 2007-11-20 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | X-Ray structure of the P64488 from E.coli (strain K12).
To be Published
|
|
8X4F
 
 | |
7K43
 
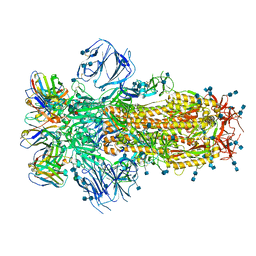 | |
3BDR
 
 | | Crystal structure of fatty acid-binding protein-like Ycf58 from Thermosynecoccus elongatus. Northeast Structural Genomics Consortium target TeR13. | | 分子名称: | PHOSPHATE ION, Ycf58 protein | | 著者 | Kuzin, A.P, Su, M, Seetharaman, J, Forouhar, F, Wang, D, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-11-15 | | 公開日 | 2007-11-27 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The crystal structure of fatty acid-binding protein-like Ycf58 from Thermosynecoccus elongatus.
To be Published
|
|
8W9W
 
 | | The cryo-EM structure of human sphingomyelin synthase-related protein in complex with ceramide/phosphoethanolamine | | 分子名称: | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, Sphingomyelin synthase-related protein 1, ~{N}-[(~{Z},2~{S},3~{R})-1,3-bis(oxidanyl)heptadec-4-en-2-yl]dodecanamide | | 著者 | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | 登録日 | 2023-09-06 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.74 Å) | | 主引用文献 | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1N5D
 
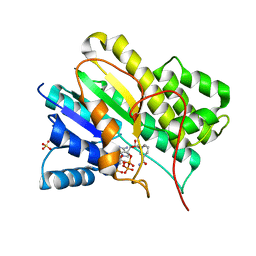 | |
3BEW
 
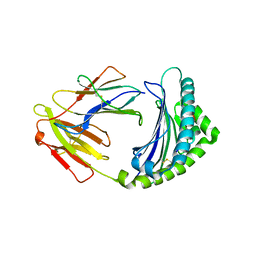 | | 10mer Crystal Structure of chicken MHC class I haplotype B21 | | 分子名称: | 10-mer from Tubulin beta-6 chain, Beta-2-microglobulin, Major histocompatibility complex class I glycoprotein haplotype B21 | | 著者 | Koch, M, Camp, S, Collen, T, Avila, D, Salomonsen, J, Wallny, H.J, van Hateren, A, Hunt, L, Jacob, J.P, Johnston, F, Marston, D.A, Shaw, I, Dunbar, P.R, Cerundolo, V, Jones, E.Y, Kaufman, J. | | 登録日 | 2007-11-20 | | 公開日 | 2008-01-01 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structures of an MHC class I molecule from b21 chickens illustrate promiscuous Peptide binding
Immunity, 27, 2007
|
|
7S1J
 
 | | Wild-type Escherichia coli ribosome with antibiotic radezolid | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | 登録日 | 2021-09-02 | | 公開日 | 2022-03-02 | | 実験手法 | ELECTRON MICROSCOPY (2.47 Å) | | 主引用文献 | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3B3C
 
 | | Crystal structure of the M180A mutant of the aminopeptidase from Vibrio proteolyticus in complex with leucine phosphonic acid | | 分子名称: | Bacterial leucyl aminopeptidase, LEUCINE PHOSPHONIC ACID, POTASSIUM ION, ... | | 著者 | Ataie, N.J, Hoang, Q.Q, Petsko, G.A, Ringe, D. | | 登録日 | 2007-10-19 | | 公開日 | 2007-11-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
7S1I
 
 | | Wild-type Escherichia coli stalled ribosome with antibiotic radezolid | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | 登録日 | 2021-09-02 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.48 Å) | | 主引用文献 | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
2W0J
 
 | | Crystal structure of Chk2 in complex with NSC 109555, a specific inhibitor | | 分子名称: | 4,4'-DIACETYLDIPHENYLUREA-BIS(GUANYLHYDRAZONE), NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | 著者 | Lountos, G.T, Tropea, J.E, Zhang, D, Jobson, A.G, Pommier, Y, Shoemaker, R.H, Waugh, D.S. | | 登録日 | 2008-08-18 | | 公開日 | 2009-02-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal Structure of Checkpoint Kinase 2 in Complex with Nsc 109555, a Potent and Selective Inhibitor
Protein Sci., 18, 2009
|
|
8X2K
 
 | | a lipid receptor complex structure | | 分子名称: | (Z)-N-(2-hydroxyethyl)octadec-9-enamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Geng, C, Yang, D, Jun, X. | | 登録日 | 2023-11-09 | | 公開日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | a lipid receptor complex structure
To Be Published
|
|
3B3S
 
 | | Crystal structure of the M180A mutant of the aminopeptidase from Vibrio proteolyticus in complex with leucine | | 分子名称: | Bacterial leucyl aminopeptidase, LEUCINE, SODIUM ION, ... | | 著者 | Ataie, N.J, Hoang, Q.Q, Petsko, G.A, Ringe, D. | | 登録日 | 2007-10-22 | | 公開日 | 2007-11-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.18 Å) | | 主引用文献 | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
3B75
 
 | | Crystal Structure of Glycated Human Haemoglobin | | 分子名称: | Hemoglobin subunit alpha, Hemoglobin subunit beta, OXYGEN MOLECULE, ... | | 著者 | Saraswathi, N.T, Syakhovich, V.E, Bokut, S.B, Moras, D, Ruff, M. | | 登録日 | 2007-10-30 | | 公開日 | 2008-10-14 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The effect of hemoglobin glycosylation on diabete linked oxidative stress
To be Published
|
|
4BVD
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | 分子名称: | 5-chloro-2-fluoro-N-[1-(4-piperidyl)pyrazol-4-yl]benzenesulfonamide, APOLIPOPROTEIN(A), CHLORIDE ION | | 著者 | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | 登録日 | 2013-06-25 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
6C9Y
 
 | | Cryo-EM structure of E. coli RNAP sigma70 holoenzyme | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Narayanan, A, Vago, F, Li, K, Qayyum, M.Z, Yenool, D, Jiang, W, Murakami, K.S. | | 登録日 | 2018-01-29 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.25 Å) | | 主引用文献 | Cryo-EM structure ofEscherichia colisigma70RNA polymerase and promoter DNA complex revealed a role of sigma non-conserved region during the open complex formation.
J. Biol. Chem., 293, 2018
|
|
2CI1
 
 | | Crystal Structure of dimethylarginine dimethylaminohydrolase I in complex with S-nitroso-Lhomocysteine | | 分子名称: | CITRIC ACID, NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1 | | 著者 | Frey, D, Braun, O, Briand, C, Vasak, M, Grutter, M.G. | | 登録日 | 2006-03-17 | | 公開日 | 2006-05-17 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | Structure of the Mammalian Nos Regulator Dimethylarginine Dimethylaminohydrolase: A Basis for the Design of Specific Inhibitors.
Structure, 14, 2006
|
|
3B9C
 
 | | Crystal Structure of Human GRP CRD | | 分子名称: | BETA-MERCAPTOETHANOL, HSPC159, SULFATE ION | | 著者 | Zhou, D, Ge, H.H, Niu, L.W, Teng, M.K. | | 登録日 | 2007-11-05 | | 公開日 | 2008-03-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the C-terminal conserved domain of human GRP, a galectin-related protein, reveals a function mode different from those of galectins.
Proteins, 71, 2008
|
|
