6Y9Z
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,9) | | 分子名称: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2020-03-10 | | 公開日 | 2020-08-19 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6YJ5
 
 | |
6Y9V
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-8,13) | | 分子名称: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2020-03-10 | | 公開日 | 2020-08-19 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Y9Y
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-7,13) | | 分子名称: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2020-03-10 | | 公開日 | 2020-08-19 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (6.1 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Y9W
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,8) | | 分子名称: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2020-03-10 | | 公開日 | 2020-08-19 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZDJ
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,10) | | 分子名称: | Gag protein, Peptidyl-prolyl cis-trans isomerase A | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2020-06-14 | | 公開日 | 2020-08-19 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (5.8 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4QO4
 
 | |
8GT6
 
 | | human STING With agonist HB3089 | | 分子名称: | 1-[(2E)-4-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-[3-(morpholin-4-yl)propoxy]-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methyl-1H-furo[3,2-e]benzimidazole-5-carboxamide, Stimulator of interferon genes protein | | 著者 | Wang, Z, Yu, X. | | 登録日 | 2022-09-07 | | 公開日 | 2022-12-28 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
8GSZ
 
 | | Structure of STING SAVI-related mutant V147L | | 分子名称: | Stimulator of interferon genes protein | | 著者 | Wang, Z, Yu, X. | | 登録日 | 2022-09-07 | | 公開日 | 2022-12-28 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
7V2T
 
 | | The complex structure of SoBcmB and its natural precursor 2 | | 分子名称: | (3S,6S)-3-((R)-2,3-dihydroxy-2-methylpropyl)-6-((S)-4-hydroxybutan-2-yl)piperazine-2,5-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | 著者 | Zhou, J.H, Wu, L. | | 登録日 | 2021-08-09 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (2.20005679 Å) | | 主引用文献 | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
8VXA
 
 | |
7V2U
 
 | | The complex structure of SoBcmB and its product 2f | | 分子名称: | (1S,5S,6S)-5-methyl-1-[(1S,2S)-2-methyl-1,2,3-tris(oxidanyl)propyl]-2-oxa-7,9-diazabicyclo[4.2.2]decane-8,10-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | 著者 | Wu, L, Zhou, J.H. | | 登録日 | 2021-08-09 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (2.00009918 Å) | | 主引用文献 | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V2X
 
 | | The complex structure of soBcmB and its substrate 1 | | 分子名称: | (3S,6S)-3-[(2S)-butan-2-yl]-6-[(2R)-2-methyl-2,3-bis(oxidanyl)propyl]piperazine-2,5-dion, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | 著者 | Wu, L, Zhou, J.H. | | 登録日 | 2021-08-10 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (2.08387423 Å) | | 主引用文献 | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
8ERR
 
 | |
8ERQ
 
 | |
7JWB
 
 | |
8VXY
 
 | | Structure of HamA(E138A,K140A)B-plasmid DNA complex from the Escherichia coli Hachiman defense system | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, HamA, HamB, ... | | 著者 | Tuck, O.T, Hu, J.J, Doudna, J.A. | | 登録日 | 2024-02-06 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.19 Å) | | 主引用文献 | Hachiman is a genome integrity sensor.
Biorxiv, 2024
|
|
8VX9
 
 | |
8VXC
 
 | |
4QOC
 
 | |
5HSM
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS MARR FAMILY PROTEIN RV2887 | | 分子名称: | SULFATE ION, Uncharacterized HTH-type transcriptional regulator Rv2887 | | 著者 | Gao, Y.R, Li, D.F, Wang, D.C, Bi, L.J. | | 登録日 | 2016-01-26 | | 公開日 | 2017-02-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural analysis of the regulatory mechanism of MarR protein Rv2887 in M. tuberculosis
Sci Rep, 7, 2017
|
|
8KCO
 
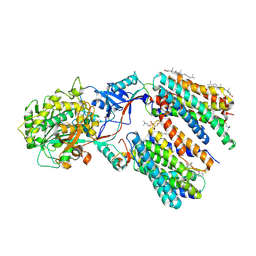 | | Cryo-EM structure of human gamma-secretase in complex with RO4929097 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2,2-dimethyl-N-[(7S)-6-oxo-5,7-dihydrobenzo[d][1]benzazepin-7-yl]-N'-(2,2,3,3,3-pentafluoropropyl)propanediamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Guo, X, Li, H, Kai, U, Yan, C, Lei, J, Zhou, R, Shi, Y. | | 登録日 | 2023-08-08 | | 公開日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | CryoEM structure of PS1-containing gamma-secretase in complex with MRK-560
To Be Published
|
|
5HSO
 
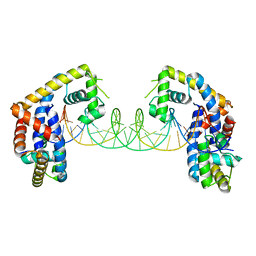 | | Crystal structure of MYCOBACTERIUM TUBERCULOSIS MARR FAMILY PROTEIN Rv2887 complex with DNA | | 分子名称: | DNA (30-MER), the upstream sequence of Rv0560c, Uncharacterized HTH-type transcriptional regulator Rv2887 | | 著者 | Gao, Y.R, Li, D.F, Wang, D.C, Bi, L.J. | | 登録日 | 2016-01-26 | | 公開日 | 2017-02-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural analysis of the regulatory mechanism of MarR protein Rv2887 in M. tuberculosis
Sci Rep, 7, 2017
|
|
6E5P
 
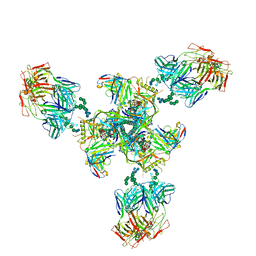 | | Backbone model based on cryo-EM map at 8.5 A of domain-swapped, glycan-reactive, neutralizing antibody 2G12 bound to HIV-1 Env BG505 DS-SOSIP, which was also bound to CD4-binding site antibody VRC03 | | 分子名称: | 2G12 Light chain, 2G12 heavy chain, Envelope glycoprotein gp120, ... | | 著者 | Acharya, P, Kwong, P.D. | | 登録日 | 2018-07-21 | | 公開日 | 2019-02-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (8.8 Å) | | 主引用文献 | Structural Survey of Broadly Neutralizing Antibodies Targeting the HIV-1 Env Trimer Delineates Epitope Categories and Characteristics of Recognition.
Structure, 27, 2019
|
|
6ENC
 
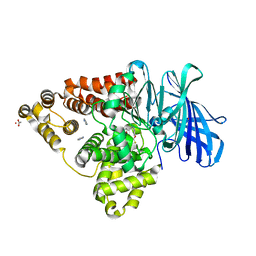 | | LTA4 hydrolase in complex with Compound11 | | 分子名称: | 1-[[4-(1,3-benzothiazol-2-yloxy)phenyl]methyl]piperidine-4-carboxylic acid, ACETATE ION, IMIDAZOLE, ... | | 著者 | Srinivas, H. | | 登録日 | 2017-10-04 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Feasibility and physiological relevance of designing highly potent aminopeptidase-sparing leukotriene A4 hydrolase inhibitors.
Sci Rep, 7, 2017
|
|
