5JS9
 
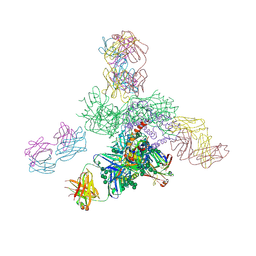 | | Uncleaved prefusion optimized gp140 trimer with an engineered 8-residue HR1 turn bound to broadly neutralizing antibodies 8ANC195 and PGT128 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Kong, L, Wilson, I.A. | | 登録日 | 2016-05-07 | | 公開日 | 2016-07-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (6.918 Å) | | 主引用文献 | Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability.
Nat Commun, 7, 2016
|
|
5JSA
 
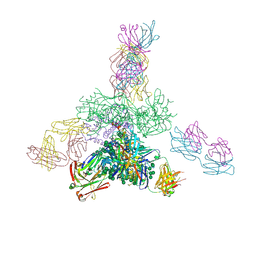 | | Uncleaved prefusion optimized gp140 trimer with an engineered 10-residue HR1 turn bound to broadly neutralizing antibodies 8ANC195 and PGT128 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Kong, L, Wilson, I.A. | | 登録日 | 2016-05-07 | | 公開日 | 2016-07-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (6.308 Å) | | 主引用文献 | Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability.
Nat Commun, 7, 2016
|
|
8IYX
 
 | | Cryo-EM structure of the GPR34 receptor in complex with the antagonist YL-365 | | 分子名称: | 1-[4-(3-chlorophenyl)phenyl]carbonyl-4-[2-(4-phenylmethoxyphenyl)ethanoylamino]piperidine-4-carboxylic acid, Probable G-protein coupled receptor 34,YL-365 | | 著者 | Jia, G.W, Wang, X, Zhang, C.B, Dong, H.H, Su, Z.M. | | 登録日 | 2023-04-06 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.34 Å) | | 主引用文献 | Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6A52
 
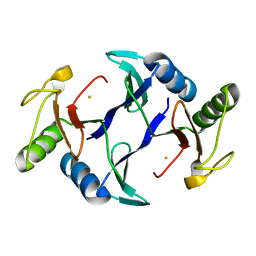 | | Oxidase ChaP-H1 | | 分子名称: | FE (II) ION, dioxidase ChaP-H1 | | 著者 | Zhang, B, Ge, H.M. | | 登録日 | 2018-06-21 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
6A5F
 
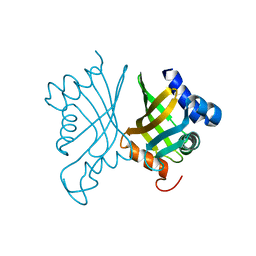 | |
6A4X
 
 | | Oxidase ChaP-H2 | | 分子名称: | Bleomycin resistance protein, FE (II) ION | | 著者 | Zhang, B, Wang, Y.S, Ge, H.M. | | 登録日 | 2018-06-21 | | 公開日 | 2018-08-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
2YEX
 
 | | Synthesis and evaluation of triazolones as checkpoint kinase 1 inhibitors | | 分子名称: | 5-METHYL-8-(1H-PYRROL-2-YL)[1,2,4]TRIAZOLO[4,3-A]QUINOLIN-1(2H)-ONE, GLYCEROL, SERINE/THREONINE-PROTEIN KINASE CHK1, ... | | 著者 | Read, J.A, Breed, J, Haye, H, McCall, E, Vallentine, A, White, A. | | 登録日 | 2011-03-31 | | 公開日 | 2012-03-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Synthesis and Evaluation of Triazolones as Checkpoint Kinase 1 Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6A4Z
 
 | | Oxidase ChaP | | 分子名称: | ChaP protein, FE (II) ION | | 著者 | Zhang, B, Ge, H.M. | | 登録日 | 2018-06-21 | | 公開日 | 2018-08-29 | | 最終更新日 | 2018-09-19 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
6A5G
 
 | |
2YER
 
 | | Synthesis and evaluation of triazolones as checkpoint kinase 1 inhibitors | | 分子名称: | 1,2-ETHANEDIOL, 5-(HYDROXYMETHYL)-8-(1H-PYRROL-2-YL)-2H-[1,2,4]TRIAZOLO[4,3-A]QUINOLIN-1-ONE, SERINE/THREONINE-PROTEIN KINASE CHK1, ... | | 著者 | Read, J.A, Breed, J, Haye, H, McCall, E, Vallentine, A, White, A, Otterbein, L. | | 登録日 | 2011-03-30 | | 公開日 | 2012-03-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Synthesis and Evaluation of Triazolones as Checkpoint Kinase 1 Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
7CUQ
 
 | | 2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | 著者 | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | 登録日 | 2020-08-24 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUW
 
 | | Ubiquinol Binding Site of Cytochrome bo3 from Escherichia coli | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | 著者 | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | 登録日 | 2020-08-25 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.63 Å) | | 主引用文献 | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUB
 
 | | 2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | 著者 | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | 登録日 | 2020-08-22 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.55 Å) | | 主引用文献 | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6A5H
 
 | |
5TBY
 
 | | HUMAN BETA CARDIAC HEAVY MEROMYOSIN INTERACTING-HEADS MOTIF OBTAINED BY HOMOLOGY MODELING (USING SWISS-MODEL) OF HUMAN SEQUENCE FROM APHONOPELMA HOMOLOGY MODEL (PDB-3JBH), RIGIDLY FITTED TO HUMAN BETA-CARDIAC NEGATIVELY STAINED THICK FILAMENT 3D-RECONSTRUCTION (EMD-2240) | | 分子名称: | Myosin light chain 3, Myosin regulatory light chain 2, ventricular/cardiac muscle isoform, ... | | 著者 | ALAMO, L, WARE, J.S, PINTO, A, GILLILAN, R.E, SEIDMAN, J.G, SEIDMAN, C.E, PADRON, R. | | 登録日 | 2016-09-13 | | 公開日 | 2017-06-07 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (20 Å) | | 主引用文献 | Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes.
Elife, 6, 2017
|
|
6CE0
 
 | | Crystal structure of a HIV-1 clade B tier-3 isolate H078.14 UFO-BG Env trimer in complex with broadly neutralizing Fabs PGT124 and 35O22 at 4.6 Angstrom | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Heavy chain, ... | | 著者 | Kumar, S, Sarkar, A, Wilson, I.A. | | 登録日 | 2018-02-09 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (4.602 Å) | | 主引用文献 | HIV-1 vaccine design through minimizing envelope metastability.
Sci Adv, 4, 2018
|
|
7F02
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | 著者 | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | 登録日 | 2021-06-03 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7F03
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with ANP | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | 著者 | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | 登録日 | 2021-06-03 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7F04
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with Heme and ATP. | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, ... | | 著者 | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | 登録日 | 2021-06-03 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
8HL9
 
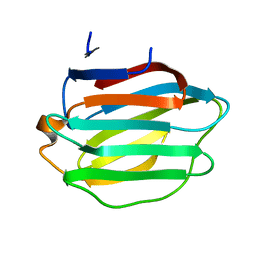 | |
7TPR
 
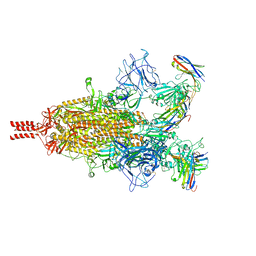 | | Camel nanobodies 7A3 and 8A2 broadly neutralize SARS-CoV-2 variants | | 分子名称: | Nanobody 7A3, Nanobody 8A2, Spike glycoprotein | | 著者 | Butay, K.J, Zhu, J, Dandey, V.P, Hong, J, Kwon, H.J, Chen, C.Z, Duan, Z, Li, D, Ren, H, Liang, T, Martin, N, Esposito, D, Ortega-Rodriguez, U, Xu, M, Xie, H, Ho, M, Cachau, R, Borgnia, M.J. | | 登録日 | 2022-01-25 | | 公開日 | 2022-04-20 | | 実験手法 | ELECTRON MICROSCOPY (2.39 Å) | | 主引用文献 | Camel nanobodies broadly neutralize SARS-CoV-2 variants
bioRxiv, 2021
|
|
5XDZ
 
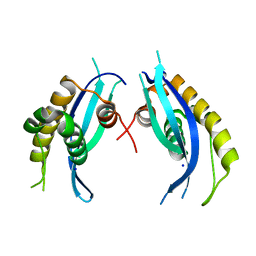 | | Crystal structure of zebrafish SNX25 PX domain | | 分子名称: | CHLORIDE ION, Cellular trafficking protein, SODIUM ION | | 著者 | Su, K, Zhang, Y, Xu, J, Liu, J. | | 登録日 | 2017-03-30 | | 公開日 | 2017-06-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of the PX domain of SNX25 reveals a novel phospholipid recognition model by dimerization in the PX domain
FEBS Lett., 591, 2017
|
|
3QW9
 
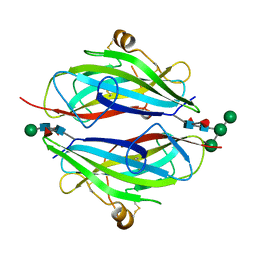 | | Crystal structure of betaglycan ZP-C domain | | 分子名称: | Transforming growth factor beta receptor type 3, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Lin, S.J, Jardetzky, T.S. | | 登録日 | 2011-02-27 | | 公開日 | 2011-04-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of betaglycan zona pellucida (ZP)-C domain provides insights into ZP-mediated protein polymerization and TGF-{beta} binding.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7WF9
 
 | |
7WF8
 
 | | Crystal structure of mouse SNX25 RGS domain in space group P212121 | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Zhang, Y, Xu, J, Liu, J. | | 登録日 | 2021-12-26 | | 公開日 | 2022-10-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structural Studies Reveal Unique Non-canonical Regulators of G Protein Signaling Homology (RH) Domains in Sorting Nexins.
J.Mol.Biol., 434, 2022
|
|
