5ONU
 
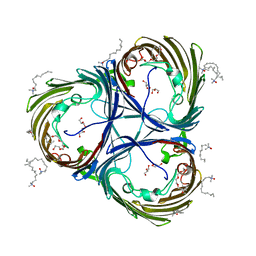 | | Trimeric OmpU structure | | 分子名称: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, GLYCEROL, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | 著者 | Li, H.Y, Dong, C.J. | | 登録日 | 2017-08-04 | | 公開日 | 2018-01-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Crystal structure of the outer membrane protein OmpU from Vibrio cholerae at 2.2 angstrom resolution.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4V68
 
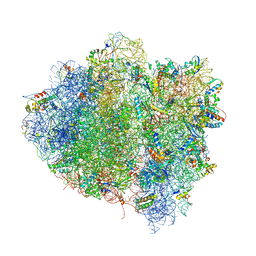 | | T. thermophilus 70S ribosome in complex with mRNA, tRNAs and EF-Tu.GDP.kirromycin ternary complex, fitted to a 6.4 A Cryo-EM map. | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Schuette, J.-C, Spahn, C.M.T. | | 登録日 | 2008-12-11 | | 公開日 | 2014-07-09 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | GTPase activation of elongation factor EF-Tu by the ribosome during decoding
Embo J., 28, 2009
|
|
4W9N
 
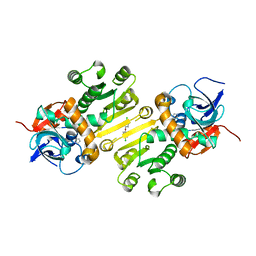 | | Enoyl-acyl carrier protein-reductase domain from human fatty acid synthase complexed with triclosan | | 分子名称: | CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase, IMIDAZOLE, ... | | 著者 | Sippel, K.H, Vyas, N.K, Sankaran, B, Quiocho, F.A. | | 登録日 | 2014-08-27 | | 公開日 | 2014-10-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Crystal structure of the human Fatty Acid synthase enoyl-acyl carrier protein-reductase domain complexed with triclosan reveals allosteric protein-protein interface inhibition.
J.Biol.Chem., 289, 2014
|
|
4W82
 
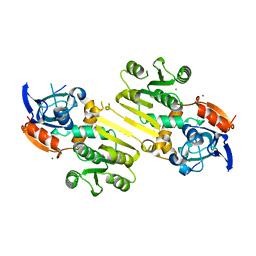 | | Enoyl-acyl carrier protein-reductase domain from human fatty acid synthase | | 分子名称: | CHLORIDE ION, Fatty acid synthase, MAGNESIUM ION, ... | | 著者 | Sippel, K.H, Vyas, N.K, Sankaran, B, Quiocho, F.A. | | 登録日 | 2014-08-22 | | 公開日 | 2014-10-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of the human Fatty Acid synthase enoyl-acyl carrier protein-reductase domain complexed with triclosan reveals allosteric protein-protein interface inhibition.
J.Biol.Chem., 289, 2014
|
|
4LN0
 
 | | Crystal structure of the VGLL4-TEAD4 complex | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Transcription cofactor vestigial-like protein 4, ... | | 著者 | Wang, H, Shi, Z, Zhou, Z. | | 登録日 | 2013-07-11 | | 公開日 | 2014-02-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.896 Å) | | 主引用文献 | A Peptide Mimicking VGLL4 Function Acts as a YAP Antagonist Therapy against Gastric Cancer.
Cancer Cell, 25, 2014
|
|
6L8Q
 
 | | Complex structure of bat CD26 and MERS-RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | 著者 | Yuan, Y. | | 登録日 | 2019-11-07 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Molecular Basis of Binding between Middle East Respiratory Syndrome Coronavirus and CD26 from Seven Bat Species.
J.Virol., 94, 2020
|
|
8Q79
 
 | | Structure of mBaoJin at pH 6.5 | | 分子名称: | CHLORIDE ION, mBaoJin | | 著者 | Samygina, V.R, Subach, O.M, Vlaskina, A.V, Nikolaeva, A.Y, Borshchevsky, V, Qin, W, Subach, F.V. | | 登録日 | 2023-08-15 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 21, 2024
|
|
4ZHY
 
 | | Crystal structure of a bacterial signalling complex | | 分子名称: | FORMIC ACID, SULFATE ION, YfiB, ... | | 著者 | Li, S, Li, T, Wang, Y, Bartlam, M. | | 登録日 | 2015-04-27 | | 公開日 | 2016-04-27 | | 実験手法 | X-RAY DIFFRACTION (1.969 Å) | | 主引用文献 | Structural insights into YfiR sequestering by YfiB in Pseudomonas aeruginosa PAO1
Sci Rep, 5, 2015
|
|
4ZHV
 
 | | Crystal structure of a bacterial signalling protein | | 分子名称: | SULFATE ION, YfiB | | 著者 | Li, S, Li, T, Wang, Y, Bartlam, M. | | 登録日 | 2015-04-27 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.585 Å) | | 主引用文献 | Structural insights into YfiR sequestering by YfiB in Pseudomonas aeruginosa PAO1
Sci Rep, 5, 2015
|
|
8QBJ
 
 | | Structure of mBaoJin at pH 4.6 | | 分子名称: | CHLORIDE ION, mBaoJin | | 著者 | Samygina, V.R, Vlaskina, A.V, Gabdulkhakov, A, Subach, O.M, Subach, F.V. | | 登録日 | 2023-08-24 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 21, 2024
|
|
8QDD
 
 | | Structure of mBaoJin at pH 8.5 | | 分子名称: | CHLORIDE ION, SULFATE ION, mBaoJin | | 著者 | Samygina, V.R, Vlaskina, A.V, Gabdulkhakov, A, Subach, O.M, Subach, F.V. | | 登録日 | 2023-08-28 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 21, 2024
|
|
4X3O
 
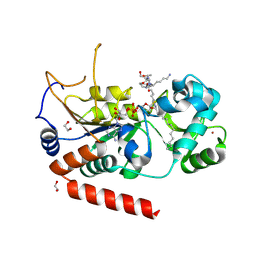 | | Sirt2 in complex with a myristoyl peptide | | 分子名称: | 1,2-ETHANEDIOL, NAD-dependent protein deacetylase sirtuin-2, ZINC ION, ... | | 著者 | Wang, Y, Zhang, W.Z, Hao, Q. | | 登録日 | 2014-12-01 | | 公開日 | 2016-01-13 | | 最終更新日 | 2022-04-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Deacylation Mechanism by SIRT2 Revealed in the 1'-SH-2'-O-Myristoyl Intermediate Structure.
Cell Chem Biol, 24, 2017
|
|
5DH3
 
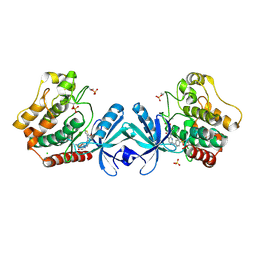 | | Crystal structure of MST2 in complex with XMU-MP-1 | | 分子名称: | 4-[(5,10-dimethyl-6-oxo-6,10-dihydro-5H-pyrimido[5,4-b]thieno[3,2-e][1,4]diazepin-2-yl)amino]benzenesulfonamide, CHLORIDE ION, SULFATE ION, ... | | 著者 | Kong, L.L, Yun, C.H. | | 登録日 | 2015-08-29 | | 公開日 | 2016-08-31 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.468 Å) | | 主引用文献 | Pharmacological targeting of kinases MST1 and MST2 augments tissue repair and regeneration
Sci Transl Med, 8, 2016
|
|
4AW5
 
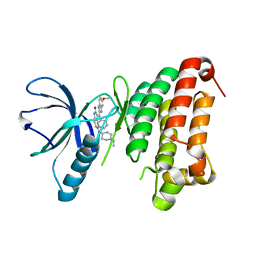 | | Complex of the EphB4 kinase domain with an oxindole inhibitor | | 分子名称: | (3Z)-5-[(1-ethylpiperidin-4-yl)amino]-3-[(5-methoxy-1H-benzimidazol-2-yl)(phenyl)methylidene]-1,3-dihydro-2H-indol-2-one, EPHRIN TYPE-B RECEPTOR 4 | | 著者 | Till, J.H, Stout, T.J. | | 登録日 | 2012-05-31 | | 公開日 | 2012-08-01 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | The Design, Synthesis, and Biological Evaluation of Potent Receptor Tyrosine Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
7CRL
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 50 ps after light activation | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-13 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRY
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (6.49 mJ/mm2) | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRK
 
 | | 2ps Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-13 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRX
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (2.63mJ/mm2) | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRT
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.17mJ/mm2) | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRS
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.90mJ/mm2) | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CU6
 
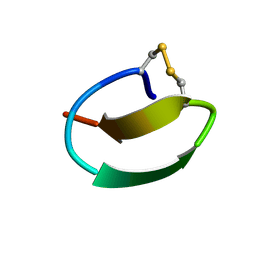 | | lasso peptide C24 mutant - A11V2C | | 分子名称: | lasso peptide C24_A11V2C | | 著者 | Ma, M, Liu, X.H. | | 登録日 | 2020-08-21 | | 公開日 | 2021-08-25 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
4FF5
 
 | |
6H35
 
 | | Myxococcus xanthus MglA bound to GDP and Pi with mixed inactive and active switch region conformations | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, GUANOSINE-5'-DIPHOSPHATE, Mutual gliding-motility protein MglA, ... | | 著者 | Varela, P.F, Galicia, C, Cherfils, J. | | 登録日 | 2018-07-17 | | 公開日 | 2019-12-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | MglA functions as a three-state GTPase to control movement reversals of Myxococcus xanthus.
Nat Commun, 10, 2019
|
|
4FZO
 
 | |
7YR5
 
 | |
