1Z1S
 
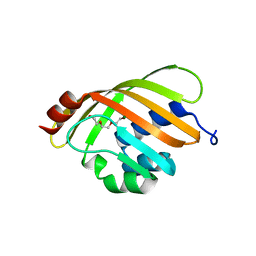 | | Crystal Structure of Putative Isomerase PA3332 from Pseudomonas aeruginosa | | 分子名称: | Hypothetical Protein PA3332, MAGNESIUM ION, TRIETHYLENE GLYCOL | | 著者 | Kim, Y, Joachimiak, A, Xu, X, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-03-06 | | 公開日 | 2005-04-19 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Crystal Structure of the Conserved Hypothetical Protein PA3332 from Pseudomonas aeruginosa
To be Published
|
|
4Q3K
 
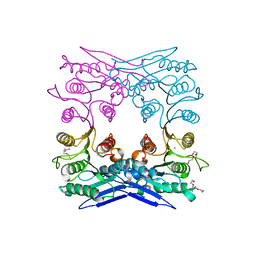 | | Crystal structure of MGS-M1, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | 分子名称: | CHLORIDE ION, FLUORIDE ION, MGS-M1, ... | | 著者 | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | 登録日 | 2014-04-11 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
2ARZ
 
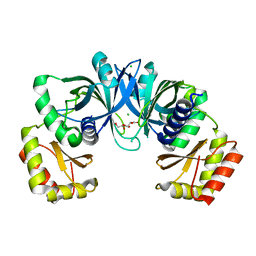 | | Crystal Structure of Protein of Unknown Function from Pseudomonas aeruginosa | | 分子名称: | CHLORIDE ION, GLYCEROL, hypothetical protein PA4388 | | 著者 | Nocek, B, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-08-22 | | 公開日 | 2005-10-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of hypothetical protein from Pseudomonas aeruginosa
TO BE PUBLISHED
|
|
4Q3L
 
 | | Crystal structure of MGS-M2, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | 分子名称: | GLYCEROL, MGS-M2 | | 著者 | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | 登録日 | 2014-04-11 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
2B0C
 
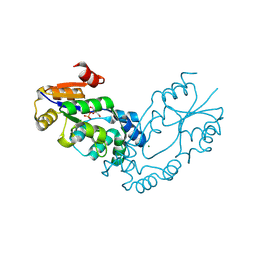 | | The crystal structure of the putative phosphatase from Escherichia coli | | 分子名称: | 1-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, putative phosphatase | | 著者 | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-09-13 | | 公開日 | 2005-11-22 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The 2.0A crystal structure of the putative phosphatase from Escherichia coli
To be Published
|
|
2B1Y
 
 | | Crystal Structure of Protein of Unknown Function ATU1913 from Agrobacterium tumefaciens str. C58 | | 分子名称: | SULFATE ION, hypothetical protein Atu1913 | | 著者 | Nocek, B, Skarina, T, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-09-16 | | 公開日 | 2005-11-01 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of Hypothetical Protein from Agrobacterium tumefaciens
reveals a new fold.
To be Published
|
|
2B3M
 
 | | Crystal structure of protein AF1124 from Archaeoglobus fulgidus | | 分子名称: | hypothetical protein AF1124 | | 著者 | Chang, C, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-09-20 | | 公開日 | 2005-11-01 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of protein AF1124 from Archaeoglobus fulgidus
To be Published
|
|
2M2J
 
 | | Solution NMR structure of the N-terminal domain of STM1478 from Salmonella typhimurium LT2: Target STR147A of the Northeast Structural Genomics consortium (NESG), and APC101565 of the Midwest Center for Structural Genomics (MCSG). | | 分子名称: | Putative periplasmic protein | | 著者 | Houliston, S, Yee, A, Lemak, A, Garcia, M, Wu, B, Savchenko, A, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-12-21 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and Functional Characterization of DUF1471 Domains of Salmonella Proteins SrfN, YdgH/SssB, and YahO.
Plos One, 9, 2014
|
|
1XSV
 
 | | X-ray crystal structure of conserved hypothetical UPF0122 protein SAV1236 from Staphylococcus aureus subsp. aureus Mu50 | | 分子名称: | Hypothetical UPF0122 protein SAV1236 | | 著者 | Walker, J.R, Xu, X, Virag, C, McDonald, M.-L, Houston, S, Buzadzija, K, Vedadi, M, Dharamsi, A, Fiebig, K.M, Savchenko, A. | | 登録日 | 2004-10-20 | | 公開日 | 2004-10-26 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | 1.7 Angstrom Crystal Structure of Conserved Hypothetical UPF0122 Protein SAV1236 From Staphylococcus aureus
To be Published
|
|
2OR0
 
 | | Structural Genomics, the crystal structure of a putative hydroxylase from Rhodococcus sp. RHA1 | | 分子名称: | ACETATE ION, Hydroxylase | | 著者 | Tan, K, Skarina, T, Kagen, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-02-01 | | 公開日 | 2007-03-06 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The crystal structure of a putative hydroxylase from Rhodococcus sp. RHA1
To be Published
|
|
3KBO
 
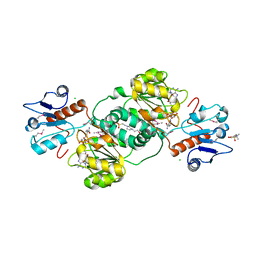 | | 2.14 Angstrom Crystal Structure of Putative Oxidoreductase (ycdW) from Salmonella typhimurium in Complex with NADP | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Glyoxylate/hydroxypyruvate reductase A, ... | | 著者 | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-10-20 | | 公開日 | 2009-10-27 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | 2.14 Angstrom Crystal Structure of Putative Oxidoreductase (ycdW) from Salmonella typhimurium in Complex with NADP.
To be Published
|
|
2OMO
 
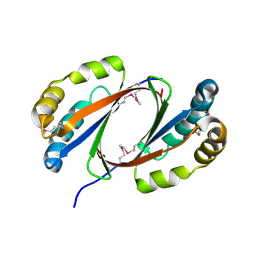 | | Putative antibiotic biosynthesis monooxygenase from Nitrosomonas europaea | | 分子名称: | DUF176 | | 著者 | Osipiuk, J, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-01-22 | | 公開日 | 2007-01-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | X-ray structure of putative antibiotic biosynthesis monooxygenase from Nitrosomonas europaea.
To be Published
|
|
3KHY
 
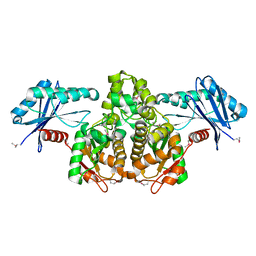 | | Crystal Structure of a propionate kinase from Francisella tularensis subsp. tularensis SCHU S4 | | 分子名称: | Propionate kinase | | 著者 | Brunzelle, J.S, Skarina, T, Sharma, S, Wang, Y, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-10-31 | | 公開日 | 2010-01-19 | | 最終更新日 | 2014-10-01 | | 実験手法 | X-RAY DIFFRACTION (1.978 Å) | | 主引用文献 | Crystal Structure of a propionate kinase from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
2OQG
 
 | | ArsR-like Transcriptional Regulator from Rhodococcus sp. RHA1 | | 分子名称: | ACETIC ACID, Possible transcriptional regulator, ArsR family protein | | 著者 | Kim, Y, Xu, X, Zheng, H, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-01-31 | | 公開日 | 2007-03-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Crystal Structure of the ArsR-like Transcriptional Regulator from Rhodococcus sp. RHA1
To be Published
|
|
5F46
 
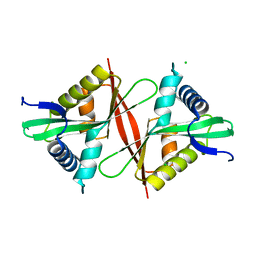 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample, apoenzyme form | | 分子名称: | CHLORIDE ION, aminoglycoside acetyltransferase meta-AAC0020 | | 著者 | Xu, Z, Skarina, T, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-12-03 | | 公開日 | 2015-12-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
5F48
 
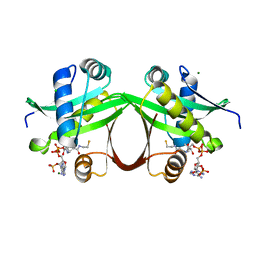 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample in complex with coenzyme A | | 分子名称: | CHLORIDE ION, COENZYME A, MAGNESIUM ION, ... | | 著者 | Xu, Z, Skarina, T, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-12-03 | | 公開日 | 2015-12-30 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
4XCW
 
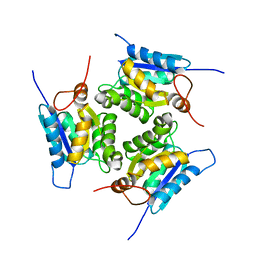 | |
5F47
 
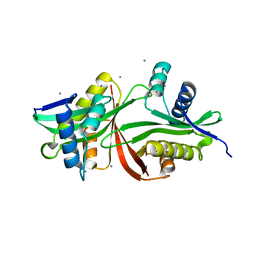 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample in complex with trehalose | | 分子名称: | CALCIUM ION, CHLORIDE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, ... | | 著者 | Xu, Z, Skarina, T, Wawrzak, Z, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-12-03 | | 公開日 | 2015-12-30 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.497 Å) | | 主引用文献 | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
3GK6
 
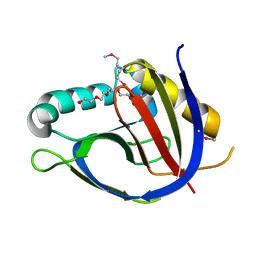 | | Crystal structure from the mobile metagenome of Vibrio cholerae. Integron cassette protein VCH_CASS2. | | 分子名称: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Integron cassette protein VCH_CASS2 | | 著者 | Deshpande, C.N, Sureshan, V, Harrop, S.J, Boucher, Y, Xu, X, Cui, H, Edwards, A.M, Savchenko, A, Joachimiak, A, Chang, C, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-03-10 | | 公開日 | 2009-03-24 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of an Integron Gene Cassette-Associated Protein from Vibrio cholerae Identifies a Cationic Drug-Binding Module.
Plos One, 6, 2011
|
|
2P12
 
 | | Crystal structure of protein of unknown function DUF402 from Rhodococcus sp. RHA1 | | 分子名称: | ACETIC ACID, GLYCEROL, Hypothetical protein DUF402 | | 著者 | Kim, Y, Evdokimova, E, Kudritska, M, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-03-01 | | 公開日 | 2007-04-03 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | The crystal structure of the protein of uncharacterized function, DUF402 from Rhodococcus sp. RHA1
To be Published
|
|
3L4E
 
 | |
2LF3
 
 | | Solution NMR structure of HopPmaL_281_385 from Pseudomonas syringae pv. maculicola str. ES4326, Midwest Center for Structural Genomics target APC40104.5 and Northeast Structural Genomics Consortium target PsT2A | | 分子名称: | Effector protein hopAB3 | | 著者 | Wu, B, Yee, A, Houliston, S, Semesi, A, Garcia, M, Singer, A.U, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | 登録日 | 2011-06-28 | | 公開日 | 2011-07-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
2PHN
 
 | | Crystal structure of an amide bond forming F420-gamma glutamyl ligase from Archaeoglobus fulgidus | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, F420-0:gamma-glutamyl ligase, ... | | 著者 | Nocek, B, Evdokimova, E, Kudritska, M, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-04-11 | | 公開日 | 2007-05-15 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structure of an Amide Bond Forming F(420):gammagamma-glutamyl Ligase from Archaeoglobus Fulgidus - A Member of a New Family of Non-ribosomal Peptide Synthases.
J.Mol.Biol., 372, 2007
|
|
2LF6
 
 | | Solution NMR structure of HopABPph1448_220_320 from Pseudomonas syringae pv. phaseolicola str. 1448A, Midwest Center for Structural Genomics target APC40132.4 and Northeast Structural Genomics Consortium target PsT3A | | 分子名称: | Effector protein hopAB1 | | 著者 | Wu, B, Yee, A, Houliston, S, Semesi, A, Garcia, M, Singer, A.U, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | 登録日 | 2011-06-28 | | 公開日 | 2011-07-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
4XV0
 
 | | Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Trichoderma reesei | | 分子名称: | Beta-xylanase, CHLORIDE ION, GLYCEROL, ... | | 著者 | Stogios, P.J, Xu, X, Cui, H, Savchenko, A. | | 登録日 | 2015-01-26 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9697 Å) | | 主引用文献 | Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Trichoderma reesei
To Be Published
|
|
