1JCI
 
 | | Stabilization of the Engineered Cation-binding Loop in Cytochrome c Peroxidase (CcP) | | 分子名称: | Cytochrome C Peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Bhaskar, B, Bonagura, C.A, Li, H, Poulos, T.L. | | 登録日 | 2001-06-09 | | 公開日 | 2002-03-06 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Cation-induced stabilization of the engineered cation-binding loop in cytochrome c peroxidase (CcP).
Biochemistry, 41, 2002
|
|
1KX5
 
 | | X-Ray Structure of the Nucleosome Core Particle, NCP147, at 1.9 A Resolution | | 分子名称: | CHLORIDE ION, DNA (5'(ATCAATATCCACCTGCAGATACTACCAAAAGTGTATTTGGAAACTGCTCCATCAAAAGGCATGTTCAGCTGGAATCCAGCTGAACATGCCTTTTGATGGAGCAGTTTCCAAATACACTTTTGGTAGTATCTGCAGGTGGATATTGAT)3'), DNA (5'(ATCAATATCCACCTGCAGATACTACCAAAAGTGTATTTGGAAACTGCTCCATCAAAAGGCATGTTCAGCTGGATTCCAGCTGAACATGCCTTTTGATGGAGCAGTTTCCAAATACACTTTTGGTAGTATCTGCAGGTGGATATTGAT)3'), ... | | 著者 | Davey, C.A, Sargent, D.F, Luger, K, Maeder, A.W, Richmond, T.J. | | 登録日 | 2002-01-31 | | 公開日 | 2002-12-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Solvent Mediated Interactions in the Structure of the Nucleosome Core Particle at 1.9 A Resolution
J.Mol.Biol., 319, 2002
|
|
3GBQ
 
 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | GRB2, SOS-1 | | 著者 | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | 登録日 | 1996-12-23 | | 公開日 | 1997-09-04 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
2GBQ
 
 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, 15 STRUCTURES | | 分子名称: | GRB2, SOS-1 | | 著者 | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | 登録日 | 1996-12-23 | | 公開日 | 1997-09-04 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
2H39
 
 | | Crystal Structure of an ADP-Glucose Phosphorylase from Arabidopsis thaliana with bound ADP-Glucose | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE-GLUCOSE, CHLORIDE ION, Probable galactose-1-phosphate uridyl transferase, ... | | 著者 | McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2006-05-22 | | 公開日 | 2006-06-13 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Crystal Structure of an ADP-Glucose Phosphorylase from Arabidopsis thaliana with bound ADP-Glucose
To be Published
|
|
7T7G
 
 | | Imipenem-OXA-23 2 minute complex | | 分子名称: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Beta-lactamase OXA-23 | | 著者 | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | 登録日 | 2021-12-15 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | C6 Hydroxymethyl-Substituted Carbapenem MA-1-206 Inhibits the Major Acinetobacter baumannii Carbapenemase OXA-23 by Impeding Deacylation.
Mbio, 13, 2022
|
|
7T7E
 
 | | MA-1-206-OXA-23 3 minute complex | | 分子名称: | (2R,4S)-2-(1,3-dihydroxypropan-2-yl)-4-{[(3R,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase OXA-23 | | 著者 | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | 登録日 | 2021-12-15 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | C6 Hydroxymethyl-Substituted Carbapenem MA-1-206 Inhibits the Major Acinetobacter baumannii Carbapenemase OXA-23 by Impeding Deacylation.
Mbio, 13, 2022
|
|
7T7D
 
 | | MA-1-206-OXA-23 30s complex | | 分子名称: | (2R,4S)-2-(1,3-dihydroxypropan-2-yl)-4-{[(3R,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase OXA-23 | | 著者 | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | 登録日 | 2021-12-15 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | C6 Hydroxymethyl-Substituted Carbapenem MA-1-206 Inhibits the Major Acinetobacter baumannii Carbapenemase OXA-23 by Impeding Deacylation.
Mbio, 13, 2022
|
|
7T7F
 
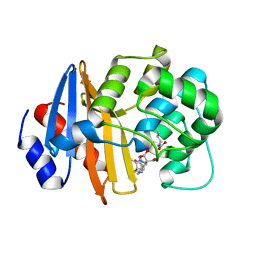 | | MA-1-206-OXA-23 25 minute complex | | 分子名称: | (2R,4S)-2-(1,3-dihydroxypropan-2-yl)-4-{[(3R,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase OXA-23 | | 著者 | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | 登録日 | 2021-12-15 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | C6 Hydroxymethyl-Substituted Carbapenem MA-1-206 Inhibits the Major Acinetobacter baumannii Carbapenemase OXA-23 by Impeding Deacylation.
Mbio, 13, 2022
|
|
7TBM
 
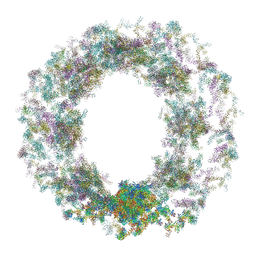 | | Composite structure of the dilated human nuclear pore complex (NPC) generated with a 37A in situ cryo-ET map of CD4+ T cell NPC | | 分子名称: | DDX19, NUP107 CTD, NUP107 NTD, ... | | 著者 | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-15 | | 最終更新日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (37 Å) | | 主引用文献 | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7TA7
 
 | |
7T9Y
 
 | |
7T8R
 
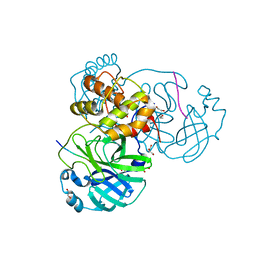 | |
7TBL
 
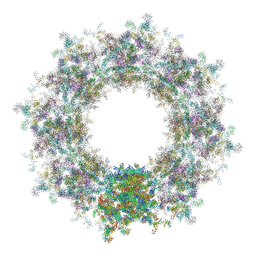 | | Composite structure of the human nuclear pore complex (NPC) cytoplasmic face generated with a 12A cryo-ET map of the purified HeLa cell NPC | | 分子名称: | DDX19, ELYS, GLE1, ... | | 著者 | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-22 | | 最終更新日 | 2022-06-29 | | 実験手法 | ELECTRON MICROSCOPY (23 Å) | | 主引用文献 | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7TC4
 
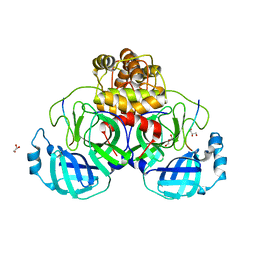 | |
7TB2
 
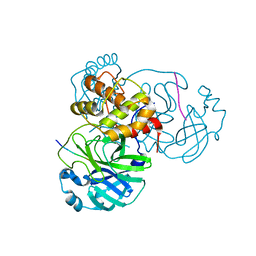 | |
7T8M
 
 | |
7T70
 
 | |
7TA4
 
 | |
7TBT
 
 | |
7TA3
 
 | |
7TA6
 
 | | Trimer-to-Monomer Disruption of Tumor Necrosis Factor-alpha (TNF-alpha) by unnatural alpha/beta-peptide-1 | | 分子名称: | 1,2-ETHANEDIOL, AMINO GROUP, Alpha/Beta-peptide-1, ... | | 著者 | Niu, J, Bingman, C.A, Gellman, S.H. | | 登録日 | 2021-12-20 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Trimer-to-Monomer Disruption Mechanism for a Potent, Protease-Resistant Antagonist of Tumor Necrosis Factor-alpha Signaling.
J.Am.Chem.Soc., 144, 2022
|
|
7SQK
 
 | | Cryo-EM structure of the human augmin complex | | 分子名称: | HAUS augmin-like complex subunit 1, HAUS augmin-like complex subunit 2, HAUS augmin-like complex subunit 3, ... | | 著者 | Gabel, C.A, Chang, L. | | 登録日 | 2021-11-05 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (8 Å) | | 主引用文献 | Molecular architecture of the augmin complex.
Nat Commun, 13, 2022
|
|
7TZ1
 
 | | Crystal Structure of a Mycobacteriophage Cluster A2 Immunity Repressor:DNA Complex | | 分子名称: | DNA (5'-D(P*CP*CP*CP*GP*CP*TP*TP*GP*AP*CP*AP*GP*CP*CP*AP*CP*CP*GP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*CP*GP*GP*TP*GP*GP*CP*TP*GP*TP*CP*AP*AP*GP*CP*GP*GP*G)-3'), Immunity repressor | | 著者 | McGinnis, R.J, Brambley, C.A, Stamey, B, Green, W.C, Gragg, K.N, Cafferty, E.R, Terwilliger, T.C, Hammel, M, Hollis, T.J, Miller, J.M, Gainey, M.D, Wallen, J.R. | | 登録日 | 2022-02-15 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-07-27 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun, 13, 2022
|
|
3HIP
 
 | |
