8AYH
 
 | |
4H6I
 
 | |
6Z3W
 
 | | Human ER membrane protein complex | | 分子名称: | ER membrane protein complex subunit 1,ER membrane protein complex subunit 1,ER membrane protein complex subunit 1,ER membrane protein complex subunit 1, ER membrane protein complex subunit 10, ER membrane protein complex subunit 2, ... | | 著者 | Hegde, R.S, O'Donnell, J.P. | | 登録日 | 2020-05-22 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | The architecture of EMC reveals a path for membrane protein insertion.
Elife, 9, 2020
|
|
7X7S
 
 | | Solution structure of human adenylate kinase 1 (hAK1) | | 分子名称: | Adenylate kinase isoenzyme 1 | | 著者 | Zhang, H. | | 登録日 | 2022-03-10 | | 公開日 | 2022-05-11 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | ADP-Induced Conformational Transition of Human Adenylate Kinase 1 Is Triggered by Suppressing Internal Motion of alpha 3 alpha 4 and alpha 7 alpha 8 Fragments on the ps-ns Timescale.
Biomolecules, 12, 2022
|
|
7OO5
 
 | |
6T6O
 
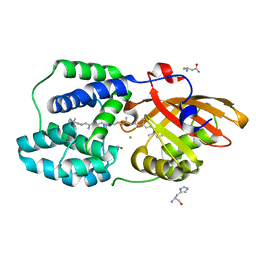 | | Y201W mutant of the orange carotenoid protein from Synechocystis at pH 4.6 | | 分子名称: | ASPARAGINE, CHLORIDE ION, GLYCEROL, ... | | 著者 | Sluchanko, N.N, Gushchin, I, Botnarevskiy, V.S, Slonimskiy, Y.B, Remeeva, A, Kovalev, K, Stepanov, A.V, Gordeliy, V, Maksimov, E.G. | | 登録日 | 2019-10-18 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Role of hydrogen bond alternation and charge transfer states in photoactivation of the Orange Carotenoid Protein.
Commun Biol, 4, 2021
|
|
8WWF
 
 | |
8WWE
 
 | |
8WWD
 
 | |
7V3S
 
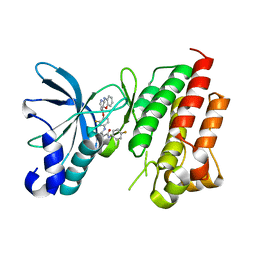 | | Crystal structure of CMET in complex with a novel inhibitor | | 分子名称: | Hepatocyte growth factor receptor, ~{N}1'-[3-fluoranyl-4-(10~{H}-pyrido[3,2-b][1,4]benzoxazin-4-yloxy)phenyl]-~{N}1-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | 著者 | Su, H.X, Liu, Q.F, Chen, T.T, Li, M.J, Xu, Y.C. | | 登録日 | 2021-08-11 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery of 10H-Benzo[b]pyrido[2,3-e][1,4]oxazine AXL Inhibitors via Structure-Based Drug Design Targeting c-Met Kinase
J.Med.Chem., 66, 2023
|
|
4K2C
 
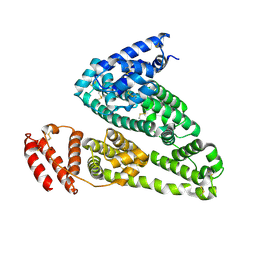 | | HSA Ligand Free | | 分子名称: | Serum albumin | | 著者 | Wang, Y, Luo, Z, Shi, X, Huang, M. | | 登録日 | 2013-04-08 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.23 Å) | | 主引用文献 | Structural mechanism of ring-opening reaction of glucose by human serum albumin.
J. Biol. Chem., 288, 2013
|
|
4IW2
 
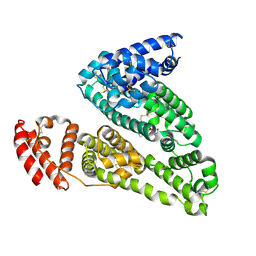 | | HSA-glucose complex | | 分子名称: | D-glucose, PHOSPHATE ION, Serum albumin, ... | | 著者 | Wang, Y, Yu, H, Shi, X, Luo, Z, Huang, M. | | 登録日 | 2013-01-23 | | 公開日 | 2013-04-24 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structural mechanism of ring-opening reaction of glucose by human serum albumin
J.Biol.Chem., 288, 2013
|
|
7V3R
 
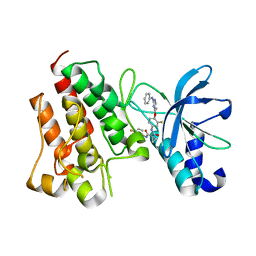 | | Crystal structure of CMET in complex with a novel inhibitor | | 分子名称: | Hepatocyte growth factor receptor, ~{N}1'-[3-fluoranyl-4-(2-phenylazanylpyrimidin-4-yl)oxy-phenyl]-~{N}1-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | 著者 | Su, H.X, Liu, Q.F, Chen, T.T, Li, M.J, Xu, Y.C. | | 登録日 | 2021-08-11 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Discovery of 10H-Benzo[b]pyrido[2,3-e][1,4]oxazine AXL Inhibitors via Structure-Based Drug Design Targeting c-Met Kinase
J.Med.Chem., 66, 2023
|
|
4IW1
 
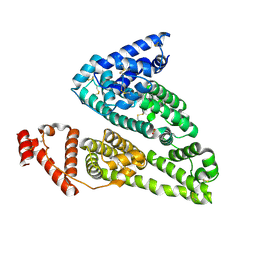 | | HSA-fructose complex | | 分子名称: | D-fructose, PHOSPHATE ION, Serum albumin, ... | | 著者 | Wang, Y, Yu, H, Shi, X, Huang, M. | | 登録日 | 2013-01-23 | | 公開日 | 2013-04-24 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Structural mechanism of ring-opening reaction of glucose by human serum albumin
J.Biol.Chem., 288, 2013
|
|
2KW0
 
 | |
1NF3
 
 | | Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6 | | 分子名称: | G25K GTP-binding protein, placental isoform, MAGNESIUM ION, ... | | 著者 | Garrard, S.M, Capaldo, C.T, Gao, L, Rosen, M.K, Macara, I.G, Tomchick, D.R. | | 登録日 | 2002-12-12 | | 公開日 | 2003-03-04 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6
Embo J., 22, 2003
|
|
2JNH
 
 | |
1JJR
 
 | |
7NTB
 
 | |
2L7W
 
 | |
6WFZ
 
 | |
6WFY
 
 | |
6PHF
 
 | |
6PHH
 
 | |
6PBW
 
 | | Crystal structure of Fab667 complex | | 分子名称: | Fab667 heavy chain, Fab667 light chain, GLYCEROL, ... | | 著者 | Oyen, D, Wilson, I.A. | | 登録日 | 2019-06-14 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.058 Å) | | 主引用文献 | Structure and mechanism of monoclonal antibody binding to the junctional epitope of Plasmodium falciparum circumsporozoite protein.
Plos Pathog., 16, 2020
|
|
