4GYY
 
 | | Crystal structure of human O-GlcNAc Transferase with UDP-5SGlcNAc and a peptide substrate | | 分子名称: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, Casein kinase II subunit alpha, SULFATE ION, ... | | 著者 | Lazarus, M.B, Jiang, J, Gloster, T.M, Zandberg, W.F, Vocadlo, D.J, Walker, S. | | 登録日 | 2012-09-05 | | 公開日 | 2012-10-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural snapshots of the reaction coordinate for O-GlcNAc transferase.
Nat.Chem.Biol., 8, 2012
|
|
4GZ6
 
 | | Crystal structure of human O-GlcNAc Transferase with UDP-5SGlcNAc | | 分子名称: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, SULFATE ION, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | 著者 | Lazarus, M.B, Jiang, J, Gloster, T.M, Zandberg, W.F, Vocadlo, D.J, Walker, S. | | 登録日 | 2012-09-06 | | 公開日 | 2012-10-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Structural snapshots of the reaction coordinate for O-GlcNAc transferase.
Nat.Chem.Biol., 8, 2012
|
|
4N39
 
 | |
4N3B
 
 | | Crystal Structure of human O-GlcNAc Transferase bound to a peptide from HCF-1 pro-repeat2(1-26)E10Q and UDP-5SGlcNAc | | 分子名称: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, Host cell factor 1, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | 著者 | Lazarus, M.B, Herr, W, Walker, S. | | 登録日 | 2013-10-06 | | 公開日 | 2014-01-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | HCF-1 is cleaved in the active site of O-GlcNAc transferase.
Science, 342, 2013
|
|
1KP7
 
 | | Conserved RNA Structure within the HCV IRES eIF3 Binding Site | | 分子名称: | Hepatitis C Virus Internal Ribosome Entry Site Fragment | | 著者 | Gallego, J, Klinck, R, Collier, A.J, Cole, P.T, Harris, S.J, Harrison, G.P, Aboul-ela, F, Walker, S, Varani, G. | | 登録日 | 2001-12-29 | | 公開日 | 2002-04-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A conserved RNA structure within the HCV IRES eIF3-binding site.
Nat.Struct.Biol., 9, 2002
|
|
4N3A
 
 | |
4N3C
 
 | |
1FQZ
 
 | | NMR VALIDATED MODEL OF DOMAIN IIID OF HEPATITIS C VIRUS INTERNAL RIBOSOME ENTRY SITE | | 分子名称: | HEPATITIS C VIRUS IRES DOMAIN IIID | | 著者 | Klinck, R, Westhof, E, Walker, S, Afshar, M, Collier, A, Aboul-ela, F. | | 登録日 | 2000-09-07 | | 公開日 | 2001-01-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A potential RNA drug target in the hepatitis C virus internal ribosomal entry site.
RNA, 6, 2000
|
|
5HGV
 
 | | Structure of an O-GlcNAc transferase point mutant, D554N in complex with peptide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, TYR-PRO-GLY-GLY-SER-THR-PRO-VAL-SER-SER-ALA-ASN-MET-MET, ... | | 著者 | Janetzko, J, Lazarus, M.B, Walker, S. | | 登録日 | 2016-01-08 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | How the glycosyltransferase OGT catalyzes amide bond cleavage.
Nat.Chem.Biol., 12, 2016
|
|
4P32
 
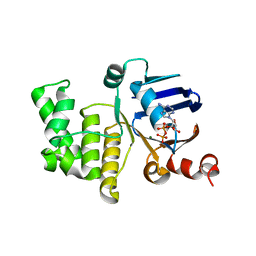 | | Crystal structure of E. coli LptB in complex with ADP-magnesium | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lipopolysaccharide export system ATP-binding protein LptB, MAGNESIUM ION | | 著者 | Sherman, D.J, Lazarus, M.B, Murphy, L, Liu, C, Walker, S, Ruiz, N, Kahne, D. | | 登録日 | 2014-03-05 | | 公開日 | 2014-03-26 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Decoupling catalytic activity from biological function of the ATPase that powers lipopolysaccharide transport.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P33
 
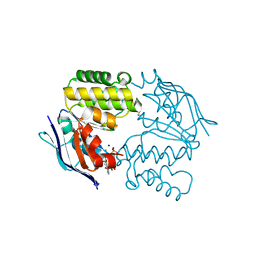 | | Crystal structure of E. coli LptB-E163Q in complex with ATP-sodium | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Lipopolysaccharide export system ATP-binding protein LptB, ... | | 著者 | Sherman, D.J, Lazarus, M.B, Murphy, L, Liu, C, Walker, S, Ruiz, N, Kahne, D. | | 登録日 | 2014-03-05 | | 公開日 | 2014-03-26 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Decoupling catalytic activity from biological function of the ATPase that powers lipopolysaccharide transport.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P31
 
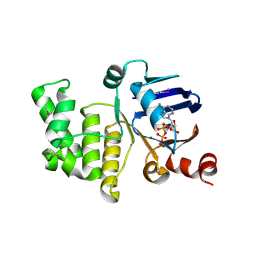 | | Crystal structure of a selenomethionine derivative of E. coli LptB in complex with ADP-Magensium | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lipopolysaccharide export system ATP-binding protein LptB, MAGNESIUM ION | | 著者 | Sherman, D.J, Lazarus, M.B, Murphy, L, Liu, C, Walker, S, Ruiz, N, Kahne, D. | | 登録日 | 2014-03-05 | | 公開日 | 2014-03-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Decoupling catalytic activity from biological function of the ATPase that powers lipopolysaccharide transport.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2OQO
 
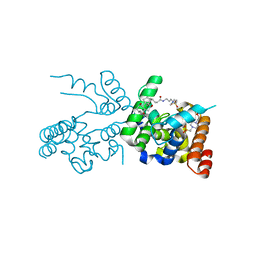 | |
4WZJ
 
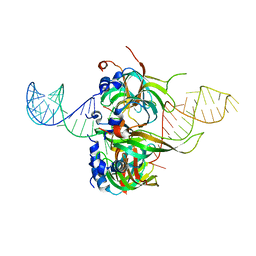 | | Spliceosomal U4 snRNP core domain | | 分子名称: | Small nuclear ribonucleoprotein E, Small nuclear ribonucleoprotein F, Small nuclear ribonucleoprotein G, ... | | 著者 | Leung, A.K.W, Nagai, K, Li, J. | | 登録日 | 2014-11-19 | | 公開日 | 2015-01-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis.
Nature, 473, 2011
|
|
3CW1
 
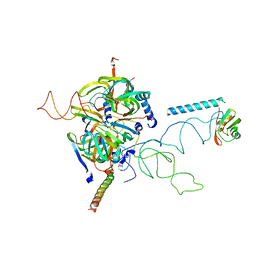 | | Crystal Structure of Human Spliceosomal U1 snRNP | | 分子名称: | Small nuclear ribonucleoprotein E, Small nuclear ribonucleoprotein F, Small nuclear ribonucleoprotein G, ... | | 著者 | Pomeranz Krummel, D.A, Oubridge, C, Leung, A.K, Li, J, Nagai, K. | | 登録日 | 2008-04-21 | | 公開日 | 2009-03-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (5.493 Å) | | 主引用文献 | Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution.
Nature, 458, 2009
|
|
8DD5
 
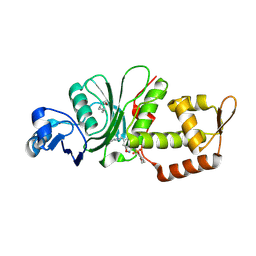 | | Crystal structure of KAT6A in complex with inhibitor CTx-648 (PF-9363) | | 分子名称: | 2,6-dimethoxy-N-{4-methoxy-6-[(1H-pyrazol-1-yl)methyl]-1,2-benzoxazol-3-yl}benzene-1-sulfonamide, Histone acetyltransferase KAT6A, ZINC ION | | 著者 | Greasley, S.E, Johnson, E, Brodsky, O. | | 登録日 | 2022-06-17 | | 公開日 | 2023-07-05 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Targeting KAT6A/KAT6B dependencies in breast cancer with a novel selective, orally bioavailable KAT6 inhibitor, CTx-648/PF-9363
To Be Published
|
|
7AUD
 
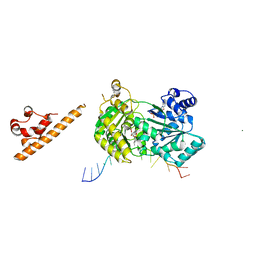 | |
7AUC
 
 | |
6PL5
 
 | | Structural coordination of polymerization and crosslinking by a peptidoglycan synthase complex | | 分子名称: | Penicillin-binding protein 2/cell division protein FtsI, Peptidoglycan glycosyltransferase RodA, Unknown peptide | | 著者 | Sjodt, M, Rohs, P.D.A, Erlandson, S.C, Zheng, S, Rudner, D.Z, Bernhardt, T.G, Kruse, A.C. | | 登録日 | 2019-06-30 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex.
Nat Microbiol, 5, 2020
|
|
6PL6
 
 | | Structural coordination of polymerization and crosslinking by a peptidoglycan synthase complex | | 分子名称: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 2/cell division protein FtsI, Peptidoglycan glycosyltransferase RodA, ... | | 著者 | Sjodt, M, Rohs, P.D.A, Erlandson, S.C, Zheng, S, Rudner, D.Z, Bernhardt, T.G, Kruse, A.C. | | 登録日 | 2019-06-30 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex.
Nat Microbiol, 5, 2020
|
|
6BAR
 
 | | Crystal structure of Thermus thermophilus Rod shape determining protein RodA (Q5SIX3_THET8) | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Rod shape determining protein RodA | | 著者 | Sjodt, M, Brock, K, Dobihal, G, Rohs, P.D.A, Green, A.G, Hopf, T.A, Meeske, A.J, Marks, D.S, Bernhardt, T.G, Rudner, D.Z, Kruse, A.C. | | 登録日 | 2017-10-15 | | 公開日 | 2018-03-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.908 Å) | | 主引用文献 | Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis.
Nature, 556, 2018
|
|
6BAS
 
 | | Crystal structure of Thermus thermophilus Rod shape determining protein RodA D255A mutant (Q5SIX3_THET8) | | 分子名称: | CHLORIDE ION, Peptidoglycan glycosyltransferase RodA | | 著者 | Sjodt, M, Brock, K, Dobihal, G, Rohs, P.D.A, Green, A.G, Hopf, T.A, Meeske, A.J, Marks, D.S, Bernhardt, T.G, Rudner, D.Z, Kruse, A.C. | | 登録日 | 2017-10-15 | | 公開日 | 2018-03-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.194 Å) | | 主引用文献 | Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis.
Nature, 556, 2018
|
|
5J3S
 
 | | Crystal structure of the catalytic domain of human tyrosyl DNA phosphodiesterase 2 in complex with a small molecule inhibitor | | 分子名称: | 2,4-dioxo-10-[3-(1H-tetrazol-5-yl)phenyl]-2,3,4,10-tetrahydropyrimido[4,5-b]quinoline-8-carbonitrile, Tyrosyl-DNA phosphodiesterase 2 | | 著者 | Hornyak, P, Pearl, L.H, Caldecott, K.W, Oliver, A.W. | | 登録日 | 2016-03-31 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Mode of action of DNA-competitive small molecule inhibitors of tyrosyl DNA phosphodiesterase 2.
Biochem.J., 473, 2016
|
|
5J3Z
 
 | | Crystal structure of m2hTDP2-CAT in complex with a small molecule inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2,4-dioxo-10-[3-(1H-tetrazol-5-yl)phenyl]-2,3,4,10-tetrahydropyrimido[4,5-b]quinoline-8-carbonitrile, ACETATE ION, ... | | 著者 | Hornyak, P, Pearl, L.H, Caldecott, K.W, Oliver, A.W. | | 登録日 | 2016-03-31 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Mode of action of DNA-competitive small molecule inhibitors of tyrosyl DNA phosphodiesterase 2.
Biochem.J., 473, 2016
|
|
5J42
 
 | | Crystal structure of m2hTDP2-CAT in complex with a small molecule inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 10-(4-hydroxyphenyl)-2,4-dioxo-2,3,4,10-tetrahydropyrimido[4,5-b]quinoline-8-carbonitrile, GLYCEROL, ... | | 著者 | Hornyak, P, Pearl, L.H, Caldecott, K.W, Oliver, A.W. | | 登録日 | 2016-03-31 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mode of action of DNA-competitive small molecule inhibitors of tyrosyl DNA phosphodiesterase 2.
Biochem.J., 473, 2016
|
|
