4JMA
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 3-Fluorocatechol | | 分子名称: | 3-FLUOROBENZENE-1,2-DIOL, Cytochrome c peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Boyce, S.E, Fischer, M, Fish, I. | | 登録日 | 2013-03-13 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JM5
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 2-Amino-5-methylthiazole | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-5-METHYLTHIAZOLE, Cytochrome c peroxidase, ... | | 著者 | Boyce, S.E, Fischer, M, Fish, I, Shoichet, B.K. | | 登録日 | 2013-03-13 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JM9
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 3-amino-1-methylpyridinium | | 分子名称: | 1-METHYL-1,6-DIHYDROPYRIDIN-3-AMINE, Cytochrome c peroxidase, IODIDE ION, ... | | 著者 | Boyce, S.E, Fischer, M, Fish, I. | | 登録日 | 2013-03-13 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JQN
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 4-Hydroxybenzaldehyde | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cytochrome c peroxidase, P-HYDROXYBENZALDEHYDE, ... | | 著者 | Boyce, S.E, Fischer, M, Fish, I. | | 登録日 | 2013-03-20 | | 公開日 | 2013-07-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JQJ
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 4-Aminoquinoline | | 分子名称: | Cytochrome c peroxidase, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Boyce, S.E, Fischer, M, Fish, I. | | 登録日 | 2013-03-20 | | 公開日 | 2013-07-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JM8
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 2,6-diaminopyridine | | 分子名称: | Cytochrome c peroxidase, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Boyce, S.E, Fischer, M, Fish, I. | | 登録日 | 2013-03-13 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
8DOA
 
 | |
6V67
 
 | | Apo Structure of the De Novo PD-1 Binding Miniprotein GR918.2 | | 分子名称: | PD-1 Binding Miniprotein GR918.2 | | 著者 | Bick, M.J, Bryan, C.M, Baker, D, Dimaio, F, Kang, A. | | 登録日 | 2019-12-04 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.07 Å) | | 主引用文献 | Computational design of a synthetic PD-1 agonist.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5VID
 
 | |
5VLI
 
 | | Computationally designed inhibitor peptide HB1.6928.2.3 in complex with influenza hemagglutinin (A/PuertoRico/8/1934) | | 分子名称: | 2,5,8,11-TETRAOXATRIDECANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Bernard, S.M, Wilson, I.A. | | 登録日 | 2017-04-25 | | 公開日 | 2017-09-27 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.799 Å) | | 主引用文献 | Massively parallel de novo protein design for targeted therapeutics.
Nature, 550, 2017
|
|
5VMR
 
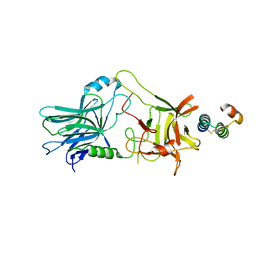 | |
5JG9
 
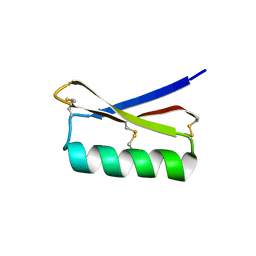 | |
5JI4
 
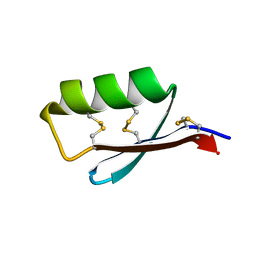 | | Solution structure of the de novo mini protein gEEHE_02 | | 分子名称: | W37 | | 著者 | Buchko, G.W, Bahl, C.D, Pulavarti, S.V, Baker, D, Szyperski, T. | | 登録日 | 2016-04-21 | | 公開日 | 2016-09-28 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
5JHI
 
 | | Solution structure of the de novo mini protein gEHE_06 | | 分子名称: | W35 | | 著者 | Buchko, G.W, Bahl, C.D, Gilmore, J.M, Pulavarti, S.V, Baker, D, Szyperski, T. | | 登録日 | 2016-04-21 | | 公開日 | 2016-09-28 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
7KUW
 
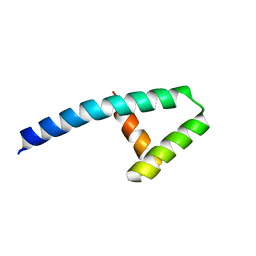 | |
3GQZ
 
 | |
3GTC
 
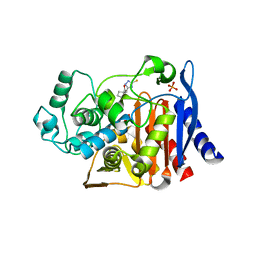 | | AmpC beta-lactamase in complex with Fragment-based Inhibitor | | 分子名称: | (1R,2S)-2-(5-thioxo-4,5-dihydro-1H-1,2,4-triazol-3-yl)cyclohexanecarboxylic acid, Beta-lactamase, PHOSPHATE ION | | 著者 | Teotico, D.T, Shoichet, B.K. | | 登録日 | 2009-03-27 | | 公開日 | 2009-04-14 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Docking for fragment inhibitors of AmpC beta-lactamase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GVB
 
 | |
3GR2
 
 | |
3GV9
 
 | |
3GRJ
 
 | |
3GSG
 
 | | AmpC beta-lactamase in complex with Fragment-based Inhibitor | | 分子名称: | (2S)-2-[(3aR,4R,7S,7aS)-1,3-dioxooctahydro-2H-4,7-methanoisoindol-2-yl]propanoic acid, Beta-lactamase, DIMETHYL SULFOXIDE, ... | | 著者 | Teotico, D.T, Shoichet, B.K. | | 登録日 | 2009-03-26 | | 公開日 | 2009-04-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Docking for fragment inhibitors of AmpC beta-lactamase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ND2
 
 | | Solution structure of the de novo mini protein gHHH_06 | | 分子名称: | De novo mini protein HHH_06 | | 著者 | Pulavarti, S.V, Eletsky, A, Bahl, C.D, Buchko, G.W, Baker, D, Szyperski, T. | | 登録日 | 2016-04-22 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
2ND3
 
 | | Solution structure of the de novo mini protein gEEH_04 | | 分子名称: | De novo mini protein EEH_04 | | 著者 | Pulavarti, S.V, Bahl, C.D, Gilmore, J.M, Eletsky, A, Buchko, G.W, Baker, D, Szyperski, T. | | 登録日 | 2016-04-22 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
5KWX
 
 | |
