7Y6O
 
 | |
7Y9A
 
 | | Crystal structure of sDscam Ig1-2 domains, isoform beta2v6 | | 分子名称: | Down Syndrome Cell Adhesion Molecules, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-3)][beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Chen, Q, Yu, Y, Cheng, J. | | 登録日 | 2022-06-24 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Structural basis for the self-recognition of sDSCAM in Chelicerata.
Nat Commun, 14, 2023
|
|
7Y4X
 
 | |
7Y5J
 
 | |
7Y5R
 
 | |
7Y73
 
 | |
7Y6E
 
 | |
7Y8S
 
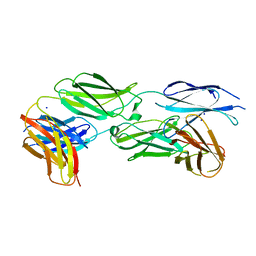 | |
7Y8I
 
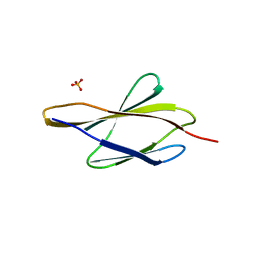 | | Crystal structure of sDscam FNIII3 domain, isoform alpha7 | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Dscam, ... | | 著者 | Chen, Q, Yu, Y, Cheng, J. | | 登録日 | 2022-06-24 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for the self-recognition of sDSCAM in Chelicerata.
Nat Commun, 14, 2023
|
|
7Y8H
 
 | |
7Y95
 
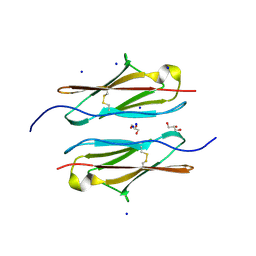 | | Crystal structure of sDscam Ig1 domain, isoform beta6v2 | | 分子名称: | Dscam, GLYCEROL, SODIUM ION | | 著者 | Chen, Q, Yu, Y, Cheng, J. | | 登録日 | 2022-06-24 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural basis for the self-recognition of sDSCAM in Chelicerata.
Nat Commun, 14, 2023
|
|
4GUS
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P3221 | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | 著者 | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | 登録日 | 2012-08-29 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
6K9V
 
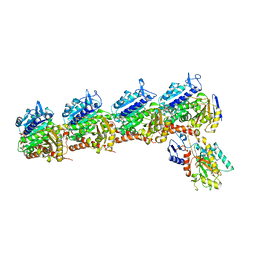 | | Crystal structure of tubulin in complex with inhibitor D64 | | 分子名称: | (5-methoxy-1H-indol-2-yl)-phenyl-methanone, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Yu, Y, Chen, Q. | | 登録日 | 2019-06-18 | | 公開日 | 2019-08-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.543 Å) | | 主引用文献 | Structural insights into the design of indole derivatives as tubulin polymerization inhibitors.
Febs Lett., 594, 2020
|
|
4GUR
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P21 | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | 著者 | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | 登録日 | 2012-08-29 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.506 Å) | | 主引用文献 | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUT
 
 | | Crystal structure of LSD2-NPAC | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1B, ... | | 著者 | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | 登録日 | 2012-08-29 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.998 Å) | | 主引用文献 | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GU1
 
 | | Crystal structure of LSD2 | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1B, ... | | 著者 | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | 登録日 | 2012-08-29 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.939 Å) | | 主引用文献 | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUU
 
 | | Crystal structure of LSD2-NPAC with tranylcypromine | | 分子名称: | Lysine-specific histone demethylase 1B, Putative oxidoreductase GLYR1, ZINC ION, ... | | 著者 | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Xu, Y. | | 登録日 | 2012-08-29 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.302 Å) | | 主引用文献 | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
7VBQ
 
 | | Heterodimer structure of Fe(II)/(alpha)ketoglutarate-dependent dioxygenase TlxIJ | | 分子名称: | FE (III) ION, Fe(II)/(alpha)ketoglutarate-dependent dioxygenase TlxI, Fe(II)/(alpha)ketoglutarate-dependent dioxygenase TlxJ, ... | | 著者 | Li, X, Awakawa, T, Mori, T, Abe, I. | | 登録日 | 2021-09-01 | | 公開日 | 2022-04-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Heterodimeric Non-heme Iron Enzymes in Fungal Meroterpenoid Biosynthesis.
J.Am.Chem.Soc., 143, 2021
|
|
7VBR
 
 | |
7C7H
 
 | |
7C7F
 
 | | Crystal structures of AKR1C3 binary complex with NADP+ | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Aldo-keto reductase family 1 member C3, ... | | 著者 | Irie, K, Toyooka, N, Endo, S. | | 登録日 | 2020-05-25 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Development of Novel AKR1C3 Inhibitors as New Potential Treatment for Castration-Resistant Prostate Cancer.
J.Med.Chem., 63, 2020
|
|
7C7G
 
 | |
8XIY
 
 | |
